Abstract
The comprehensive characterization of neuronal morphology requires tracing extensive axonal and dendritic arbors imaged with light microscopy into digital reconstructions. Considerable effort is ongoing to automate this greatly labor-intensive and currently rate-determining process. Experimental data in the form of manually traced digital reconstructions and corresponding image stacks play a vital role in developing increasingly more powerful reconstruction algorithms. The DIADEM challenge (short for DIgital reconstruction of Axonal and DEndritic Morphology) successfully stimulated progress in this area by utilizing six data set collections from different animal species, brain regions, neuron types, and visualization methods. The original research projects that provided these data are representative of the diverse scientific questions addressed in this field. At the same time, these data provide a benchmark for the types of demands automated software must meet to achieve the quality of manual reconstructions while minimizing human involvement. The DIADEM data underwent extensive curation, including quality control, metadata annotation, and format standardization, to focus the challenge on the most substantial technical obstacles. This data set package is now freely released (http://diademchallenge.org) to train, test, and aid development of automated reconstruction algorithms.








Similar content being viewed by others
References
Ascoli, G. A. (2008). Neuroinformatics grand challenges. Neuroinformatics, 6(1), 1–3.
Ascoli, G. A., Brown, K. M., Calixto, E., Card, J. P., Galván, E. J., Perez-Rosello, T., et al. (2009). Quantitative morphometry of electrophysiologically identified CA3b interneurons reveals robust local geometry and distinct cell classes. The Journal of Comparative Neurology, 515(6), 677–695.
Brown, K. M., Donohue, D. E., D’Alessandro, G., & Ascoli, G. A. (2005). A cross-platform freeware tool for digital reconstruction of neuronal arborizations from image stacks. Neuroinformatics, 3(4), 343–360.
Buckmaster, P. S., & Dudek, F. E. (1999). In vivo intracellular analysis of granule cell axon reorganization in epileptic rats. Journal of Neurophysiology, 81(2), 712–721.
Calixto, E., Galván, E. J., Card, J. P., & Barrionuevo, G. (2008). Coincidence detection of convergent perforant path and mossy fibre inputs by CA3 interneurons. The Journal of Physiology, 586(Pt 11), 2695–2712.
Canty, A. J., & De Paola, V. (2011). Axonal reconstructions going live. Neuroinformatics. doi:10.1007/s12021-011-9112-3.
De Paola, V., Arber, S., & Caroni, P. (2003). AMPA receptors regulate dynamic equilibrium of presynaptic terminals in mature hippocampal networks. Nature Neuroscience, 6(5), 491–500.
De Paola, V., Holtmaat, A., Knott, G., Song, S., Wilbrecht, L., Caroni, P., et al. (2006). Cell type-specific structural plasticity of axonal branches and boutons in the adult neocortex. Neuron, 49(6), 861–875.
Donohue, D. E., & Ascoli, G. A. (2011). Automated reconstruction of neuronal morphology: an overview. Under review upon invitation, Brain Research Reviews.
Evers, J. F., Schmitt, S., Sibila, M., & Duch, C. (2005). Progress in functional neuroanatomy: precise automatic geometric reconstruction of neuronal morphology from confocal image stacks. Journal of Neurophysiology, 93(4), 2331–2342.
Fares, T., & Stepanyants, A. (2009). Cooperative synapse formation in the neocortex. Proceedings of the National Academy of Sciences, 106(38), 16463–16468.
Feng, G., Mellor, R. H., Bernstein, M., Keller-Peck, C., Nguyen, Q. T., Wallace, M., Nerbonne, J. M., Lichtman, J. W., & Sanes, J. R. (2000). Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron, 28(1), 41–51.
Fiala, J. C. (2005). Reconstruct: a free editor for serial section microscopy. Journal of Microscopy, 218(Pt 1), 52–61.
Gao, Q., Yuan, B., & Chess, A. (2000). Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nature Neuroscience, 3(8), 780–785.
Gilbert, C. D. (1983). Microcircuitry of the visual cortex. Annual Review of Neuroscience, 6, 217–247.
Gillette, T. A., Brown, K. M., & Ascoli, G. A. (2011). The DIADEM metric: comparing multiple reconstructions of the same neuron. Neuroinformatics. doi:10.1007/s12021-011-9117-y.
Halavi, M., Polavaram, S., Donohue, D. E., Hamilton, G., Hoyt, J., Smith, K. P., et al. (2008). NeuroMorpho.Org implementation of digital neuroscience: dense coverage and integration with the NIF. Neuroinformatics, 6(3), 241–252.
Henneman, E., Somjen, G., & Carpenter, D. (1965). Functional significance of cell size in spinal motoneurons. Journal of Neurophysiology, 28(3), 560–580.
Hirsch, J. A., Alonso, J. M., Reid, R. C., & Martinez, L. M. (1998a). Synaptic integration in striate cortical simple cells. The Journal of Neuroscience, 18(22), 9517–9528.
Hirsch, J. A., Gallagher, C. A., Alonso, J. M., & Martinez, L. M. (1998b). Ascending projections of simple and complex cells in layer 6 of the cat striate cortex. The Journal of Neuroscience, 18(19), 8086–8094.
Hirsch, J. A., Martinez, L. M., Pillai, C., Alonso, J. M., Wang, Q., & Sommer, F. T. (2003). Functionally distinct inhibitory neurons at the first stage of visual cortical processing. Nature Neuroscience, 6(12), 1300–1308.
Holtmaat, A., Bonhoeffer, T., Chow, D. K., Chuckowree, J., De Paola, V., Hofer, S. B., et al. (2009). Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nature Protocols, 4(8), 1128–1144.
Jefferis, G. S. X. E., Potter, C. J., Chan, A. M., Marin, E. C., Rohlfing, T., Maurer, C. R., Jr., et al. (2007). Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell, 128(6), 1187–1203.
Kaspirzhny, A. V., Gogan, P., Horcholle-Bossavit, G., & Tyc-Dumont, S. (2002). Neuronal morphology data bases: morphological noise and assessment of data quality. Network: Computation in Neural Systems, 13, 357–380.
Lee, T., & Luo, L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron, 22(3), 451–461.
Lichtman, J. W., Livet, J., & Sanes, J. R. (2008). A technicolour approach to the connectome. Nature Reviews Neuroscience, 9, 417–422.
Livet, J., Weissman, T. A., Kang, H., Draft, R. W., Lu, J., Bennis, R. A., et al. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature, 450, 56–62.
Losavio, B. E., Liang, Y., Santamaria-Pang, A., Kakadiaris, I. A., Colbert, C. M., & Saggau, P. (2008). Live neuron morphology automatically reconstructed from multiphoton and confocal imaging data. Journal of Neurophysiology, 100, 2422–2429.
Lu, J. (2011). Neuronal tracing for connectomic studies. Neuroinformatics. doi:10.1007/s12021-011-9101-6.
Lu, J., Fiala, J. C., & Lichtman, J. W. (2009a). Semi-automated reconstruction of neural processes from large numbers of fluorescence images. PLoS ONE, 4(5), e5655.
Lu, J., Tapia, J. C., White, O. L., & Lichtman, J. W. (2009b). The interscutularis muscle connectome. PLoS Biology, 7(2), e32. Erratum in: PLoS Biology, 7(4), e1000108.
Marin, E. C., Jefferis, G. S. X. E., Komiyama, T., Zhu, H., & Luo, L. (2002). Representation of the glomerular olfactory map in the Drosophila brain. Cell, 109(2), 243–255.
Martinez, L. M., Alonso, J. M., Reid, R. C., & Hirsch, J. A. (2002). Laminar processing of stimulus orientation in cat visual cortex. The Journal of Physiology, 540(Pt 1), 321–333.
Martinez, L. M., Wang, Q., Reid, R. C., Pillai, C., Alonso, J. M., Sommer, F. T., et al. (2005). Receptive field structure varies with layer in the primary visual cortex. Nature Neuroscience, 8(3), 372–379.
Meijering, E., Jacob, M., Sarria, J. C. F., Steiner, P., Hirling, H., & Unser, M. (2004). Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry, 58A(2), 167–176.
Peng, H., Ruan, Z., Atasoy, D., & Sternson, S. (2010). Automatic reconstruction of 3D neuron structures using a graph-augmented deformable model. Bioinformatics, 26(12), i38–i46.
Rodriguez, A., Ehlenberger, D. B., Hof, P. R., & Wearne, S. L. (2009). Three-dimensional neuron tracing by voxel scooping. Journal of Neuroscience Methods, 184(1), 169–175.
Scorcioni, R., Lazarewicz, M. T., & Ascoli, G. A. (2004). Quantitative morphometry of hippocampal pyramidal cells: differences between anatomical classes and reconstructing laboratories. The Journal of Comparative Neurology, 473(2), 177–193.
Scorcioni, R., Polavaram, S., & Ascoli, G. A. (2008). L-Measure: a web-accessible tool for the analysis, comparison and search of digital reconstructions of neuronal morphologies. Nature Protocols, 3(5), 866–876.
Snider, J., Pillaim, A., & Stevens, C. F. (2010). A universal property of axonal and dendritic arbors. Neuron, 66(1), 45–56.
Sugihara, I. (2011). Bright field neuronal preparation optimized for automatic computerized reconstruction, a case with cerebellar climbing fibers. Neuroinformatics. doi:10.1007/s12021-011-9099-9.
Sugihara, I., Wu, H., & Shinoda, Y. (1999). Morphology of single olivocerebellar axons labeled with biotinylated dextran amine in the rat. The Journal of Comparative Neurology, 414(2), 131–148.
Torben-Nielsen, B., & Stiefel, K. M. (2009). Systematic mapping between dendritic function and structure. Network, 20(2), 69–105.
Volman, V., Levine, H., Ben-Jacob, E., & Sejnowski, T. J. (2009). Locally balanced dendritic integration by short-term synaptic plasticity and active dendritic conductances. Journal of Neurophysiology, 102(6), 3234–3250.
Vosshall, L. B., Wong, A. M., & Axel, R. (2000). An Olfactory Sensory Map in the Fly Brain. Cell, 102(2), 147–159.
Wittner, L., Henze, D. A., Záborszky, L., & Buzsáki, G. (2007). Three-dimensional reconstruction of the axon arbor of a CA3 pyramidal cell recorded and filled in vivo. Brain Structure and Function, 212(1), 75–83.
Information Sharing Statement
The DIADEM data sets are freely available at http://diademchallenge.org.
Acknowledgements
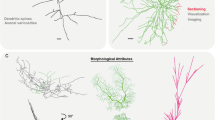
The corresponding author’s lab is supported in part by NIH R01 grant 39600. Todd A. Gillette is acknowledged for help and advice throughout the DIADEM data set organization process. Figure 4 illustration was created with V3D (http://penglab.janelia.org/proj/v3d).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Brown, K.M., Barrionuevo, G., Canty, A.J. et al. The DIADEM Data Sets: Representative Light Microscopy Images of Neuronal Morphology to Advance Automation of Digital Reconstructions. Neuroinform 9, 143–157 (2011). https://doi.org/10.1007/s12021-010-9095-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-010-9095-5