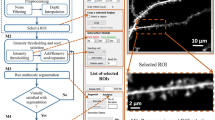
Abstract
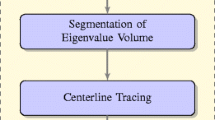
This paper presents a method for improved automatic delineation of dendrites and spines from three-dimensional (3-D) images of neurons acquired by confocal or multi-photon fluorescence microscopy. The core advance presented here is a direct grayscale skeletonization algorithm that is constrained by a structural complexity penalty using the minimum description length (MDL) principle, and additional neuroanatomy-specific constraints. The 3-D skeleton is extracted directly from the grayscale image data, avoiding errors introduced by image binarization. The MDL method achieves a practical tradeoff between the complexity of the skeleton and its coverage of the fluorescence signal. Additional advances include the use of 3-D spline smoothing of dendrites to improve spine detection, and graph-theoretic algorithms to explore and extract the dendritic structure from the grayscale skeleton using an intensity-weighted minimum spanning tree (IW-MST) algorithm. This algorithm was evaluated on 30 datasets organized in 8 groups from multiple laboratories. Spines were detected with false negative rates less than 10% on most datasets (the average is 7.1%), and the average false positive rate was 11.8%. The software is available in open source form.







Similar content being viewed by others
References
Abdul-Karim, M. A. (2005). Automated parameter selection for segmentation of tube-like biological structures using optimization algorithm and MDL. 2005 Ph.D. dissertation, Rensselaer Polytechnic Institute, Troy, NY 12180.
Abdul-Karim, M. A., Al-Kofahi, K., Brown, E. B., Jain, R. K., & Roysam, B. (2003). Automated tracing and change analysis of angiogenic vasculature from in vivo multiphoton confocal image time series. Microvascular Research, 66(2), 113–125.
Abdul-Karim, M. A., Roysam, B., Dowell-Mesfin, N. M., Jeromin, A., Yuksel, M., & Kalyanaraman, S. (2005). Automatic selection of parameters for vessel/neurite segmentation algorithms. IEEE Transactions on Image Processing, 14(9), 1338–1350.
Al-Kofahi, K. A., Can, A., Lasek, S., Szarowski, D. H., Dowell-Mesfin, N., Shain, W., et al. (2003). Median-based robust algorithms for tracing neurons from noisy confocal microscope images. IEEE Transactions on Information Technology in Biomedicine, 7(4), 302–317.
Al-Kofahi, Y., Dowell-Mesfin, N., Pace, C., Shain, W., Turner, J. N., & Roysam, B. (2008). Improved detection of branching points in algorithms for automated neuron tracing from 3-D confocal images. Cytometry A, 73(1), 36–43.
Al-Kofahi, K. A., Lasek, S., Szarowski, D. H., Pace, C. J., Nagy, G., Turner, J. N., et al. (2002). Rapid automated three-dimensional tracing of neurons from confocal image stacks. IEEE Transactions on Information Technology in Biomedicine, 6(2), 171–187.
Bai, W., Zhou, X., Ji, L., Cheng, J., & Wong, S. T. (2007). Automatic dendritic spine analysis in two-photon laser scanning microscopy images. Cytometry A, 71(10), 818–826.
Barron, A., Rissanen, J., & Yu, B. (1998). The minimum description length principle in coding and modeling. IEEE Transactions on Information Theory, 44(6), 2743–2760.
Blake, A., & Zisserman, A. (1987). Visual reconstruction. Cambridge: MIT.
Bondy, J. A., & Murty, U. S. R. (1976). Graph theory with applications. New York: Elsevier Science.
Bouman, C., & Sauer, K. (1993). A generalized Gaussian image model for edge-preserving MAP estimation. IEEE Transactions on Image Processing, 2(3), 296–310.
Cai, H., Xu, X., Lu, J., Lichtman, J. W., Yung, S. P., & Wong, S. T. (2006). Repulsive force based snake model to segment and track neuronal axons in 3-D microscopy image stacks. Neuroimage, 32(4), 1608–1620.
Cajal, S. R. Y. (1888). Estructura de los centros nervioso de las aves. Rev Trim Hitol norm Pat, 1, 1–10.
Cajal, S. R. Y. (1891). Sur la structure de l’ecorce cerebrale de quelques mammiferes. Cellule, 7, 123–176.
Can, A., Shen, H., Turner, J. N., Tanenbaum, H. L., & Roysam, B. (1999). Rapid automated tracing and feature extraction from live high-resolution retinal fundus images using direct exploratory algorithms. IEEE Transactions on Information Technology in Biomedicine, 3(2), 125–138.
Capowski, J. J. (ed). (1989). Computer techniques in neuroanatomy. New York: Plenum.
Carlsson, K., Wallen, P., & Brodin, L. (1989). Three-dimensional imaging of neurons by confocal fluorescence microscopy. Journal de Microscopie, 155(Pt 1), 15–26.
Cesar, R. M., Jr., & Costa, L. F. (1999). Semi-automated dendrogram generation for neural shape analysis. Journal of Neuroscience Methods, 93, 121–131.
Cham, T. J., & Cipolla, R. (1999). Automated B-Spline curve representation incorporating MDL and error-minimizing control point insertion strategies. IEEE Transactions on Pattern analysis and Machine Intelligence, 21(1), 49–53.
Cheng, J., Zhou, X., Miller, E., Witt, R. M., Zhu, J., Sabatini, B. L., et al. (2007a). A novel computational approach for automatic dendrite spines detection in two-photon laser scan microscopy. Journal of Neuroscience Methods, 165(1), 122–134.
Cheng, J., Zhou, X., Sabatini, B. L., & Wong, S. T. (2007b). NeuronIQ: a novel computational approach for automatic dendrite splines detection and analysis. IEEE/NIH Life Science Systems and Applications Workshop (LISSA 2007) pp. 168–71.
Cohen, A. R., Roysam, B., & Turner, J. N. (1994). Automated tracing and volume measurements of neurons from 3-D confocal fluorescence microscopy data. Journal de Microscopie, 173(Pt 2), 103–114.
Cormen, T. H., Leiserson, C. E., Rivest, R. L., & Stein, C. (2001). Introduction to algorithms (2nd ed., pp. 595–601). Cambridge: MIT.
Cornea, N. D., Silver, D., Yuan, X., & Balasubramanian, R. (2005). Computing hierarchical curve-skeletons of 3-D objects. The Visual Computer, 21(11), 945–955.
Costa, L. Da F., Manoel, E. T., Faucereau, F., Chelly, J., van Pelt, J., & Ramakers, G. (2002). Shape analysis framework for neuromorphometry. Network, 13(3), 283–310.
Dierckx, P. (1993). Curve and surface fitting with splines. Oxford: Clarendon.
Dowell-Mesfin, N. M., Abdul-Karim, M. A., Turner, A. M., Schanz, S., Craighead, H. G., Roysam, B., et al. (2004). Topographically modified surfaces affect orientation and growth of hippocampal neurons. Journal of Neural Engineering, 1(2), 78–90.
Falcao, A. X., Costa, L. F., & da Cunha, B. S. (2002). Multiscale skeletons by image foresting transform and its application to neuromorphometry. Pattern Recognition, 35(7), 1571–1582.
Frangi, A. F., Niessen, W. J., Hoogeveen, R. M., Walsum, T. V., & Viergever, M. A. (1999). Model-based quantitation of 3-D magnetic resonance angiographic images. IEEE Transactions on Medical Imaging, 18(10), 946–956.
Frangi, A. F., Niessen, W. J., Vincken, K. L., & Viergever, M. A. (1998). Multiscale vessel enhancement filtering. Medical Image Computing and Computer-Assisted Intervention, 1496, 130–137.
Glaser, E. M., Tagamets, M., McMullen, N. T., & Van der Loos, H. (1983). The image-combining computer microscope—an interactive instrument for morphometry of the nervous system. Journal of Neuroscience Methods, 8(1), 17–32.
Globus, A., Levit, C., & Lasinski, T. (1991) A tool for visualizing the topology of three-dimensional vector fields. IEEE Visualization, 33–40
Grunwald, P., Myung, J., & Pitt, M. (2004). Advances in minimum description length: Theory and applications. Cambridge: MIT Press.
Guéziec, A., & Ayache, N. (1994). Smoothing and matching of 3-D space curves. International Journal of Computer Vision, 12(1), 79–104.
Gulledge, A. T., Kampa, B. M., & Stuart, G. J. (2005). Synaptic integration in dendritic trees. Journal of Neurobiology, 64, 75–90.
He, W., Hamilton, T. A., Cohen, A. R., Holmes, T. J., Pace, C., Szarowski, D. H., et al. (2003). Automated three-dimensional tracing of neurons in confocal and brightfield images. Microscopy and Microanalysis, 9(4), 296–310.
Herzog, A., Krell, G., Michaelis, B., Wang, J., Zuschratter, W., & Braun, K. (1997). Restoration of three-dimensional quasi-binary images from confocal microscopy and its application to dendritic trees. BiOS. San Jose, 8–14.
Holmes, T. J., Bhattacharyya, S., Cooper, J. A., Hanzel, D., Krishnamurthi, V., Lin, W., et al. (1995). Light microscopic images reconstructed by maximum likelihood deconvolution. In J. Pawley (Ed.), Handbook of confocal microscopy. New York: Plenum.
Janoos, F., Nouansengsy, B., Xu, X., MacHiraju, R., & Wong, S. T. C. (2008). Classification and uncertainty visualization of dendritic spines from optical microscopy imaging. Computer Graphics Forum, 27(3), 879–886.
Kalus, P., Muller, T. J., Zuschratter, W., & Senitz, D. (2000). The dendritic architecture of prefrontal pyramidal neurons in schizophrenic patients. NeuroReport, 11(16), 3621–3625.
Kirbas, C., & Quek, F. (2004). A review of vessel extraction techniques and algorithms. ACM Computing Surveys, 36(2), 81–121.
Koh, I. Y., Lindquist, W. B., Zito, K., Nimchinsky, E. A., & Svoboda, K. (2002). An image analysis algorithm for dendritic spines. Neural Computation, 14(6), 1283–1310.
Leclerc, Y. G. (1989). Constructing simple stable descriptions for image partitioning. International Journal of Computer Vision, 3(1), 73–102.
Lippman, J., & Dunaevsky, A. (2005). Dendritic spine morphogenesis and plasticity. Journal of Neurobiology, 64(1), 47–57.
Lolive, D., Barbot, N., & Boeffard, O. (2006). Melodic contour estimation with B-spline models using a MDL criterion. Proceedings of the 11th International Conference on Speech and Computer (SPECOM) (pp. 333-338). Saint Petersburg, Russia
London, M., & Hausser, M. (2005). Dendritic computation. Annual Review of Neuroscience, 28, 503–532.
Losavio, B. E., Liang, Y., Santamaria-Pang, A., Kakadiaris, I. A., Colbert, C. M., & Saggau, P. (2008). Live neuron morphology automatically reconstructed from multiphoton and confocal imaging data. Journal of Neurophysiology, 100, 2422–2429.
Lu, F., & Milios, E. (1994). Optimal spline fitting to planar shape. Signal Processing, 37, 129–140.
Matsuzaki, M. (2007). Factors critical for the plasticity of dendritic spines and memory storage. Neuroscience Research, 57, 1–9.
Meijering, E., Jacob, M., Sarria, J. C., Steiner, Pl, Hirling, H., & Unser, M. (2004). Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry, 58A(2), 167–176.
Mel, B. W. (1994). Information-processing in dendritic trees. Neural Computation, 6, 1031–1085.
Miller, M. I., Roysam, B., Smith, K. R., & O'Sullivan, J. A. (1991). Representing and computing regular languages on massively parallel networks. IEEE Transactions on Neural Networks, 2(1), 56–72.
Pawley, J. B. (2006). Handbook of biological confocal microscopy (3rd ed.) Springer.
Perona, P., & Malik, J. (1990). Scale-space and edge detection using anisotropic diffusion. IEEE Transactions on Pattern Analysis and Machine Intelligence, 12(7), 629–639.
Potter, S. M. (1996). Vital imaging: two photons are better than one. Current Biology, 6(12), 1595–1598.
Potter, S. M. (2005). Two-photon microscopy for 4D imaging of living neurons. In R. Yuste & A. Konnerth (Eds.), Imaging in neuroscience and development: A laboratory manual (pp. 59–70). Cold Spring Harbor: Cold Spring Harbor Laboratory.
Rissanen, J. (1978). Modeling by shortest data description. Automatica, 14(5), 465–471.
Rodriguez, A., Ehlenberger, D. B., Hof, P. R., & Wearne, S. L. (2006). Rayburst sampling. An algorithm for automated three-dimensional shape analysis from laser scanning microscopy images. National Protocol, 1(4), 2152–2161.
Rodriguez, A., Ehlenberger, D., Kelliher, K., Einstein, M., Henderson, S. C., Morrison, J. H., et al. (2003). Automated reconstruction of three-dimensional neuronal morphology from laser scanning microscopy images. Methods, 30(1), 94–105.
Rogers, D. F. (1998). Procedural elements for computer graphics. Boston: McGraw-Hill.
Rolston, J. D., Wagenaar, D. A., & Potter, S. M. (2007). Precisely timed spatiotemporal patterns of neural activity in dissociated cortical cultures. Neuroscience, 148(1), 294–303.
Satou, K., Aoki, Y., Mataga, N., Hensh, T. K., & Taki, K. (2005). Automatic analysis for neuron by confocal laser scanning microscope. Optomechatronic Machine Vision, Proceedings of SPIE, 6051.
Schmitt, S., Evers, J. F., Duch, C., Scholz, M., & Obermayer, K. (2004). New methods for the computer-assisted 3-D reconstruction of neurons from confocal image stacks. Neuroimage, 23(4), 1283–1298.
Schroeder, W., Martin, K., & Lorensen, B. (1998). The visualization toolkit: An object oriented approach to 3-D graphics. Printice-Hall Inc.
Schumaker, L. L. (1981). Spine functions: Basic theory. New York: Wiley.
Scorcioni, R., Polavaram, S., & Ascoli, G. A. (2008). L-Measure: a web-accessible tool for the analysis, comparison and search of digital reconstructions of neuronal morphologies. Nature Protocols, 3(5), 866–876.
Srinivasan, R., Zhou, X., Miller, E., Lu, J., Litchman, J., & Wong, S. T. (2007). Automated axon tracking of 3-D confocal laser scanning microscopy images using guided probabilistic region merging. Neuroinformatics, 5(3), 189–203.
Theisel, H., & Weinkauf, T. (2002). Vector field metrics based on distance measures of first order critical points. Journal of WSCG, 10(3).
Trachtenberg, J. T., Chen, B. E., Knott, G. W., Feng, G., Sanes, J. R., Welker, E., et al. (2002). Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature, 420(6917), 788–794.
Turner, J. N., Szarowski, D. H., Smith, K. L., Marko, M., Leith, A., & Swann, J. W. (1991). Confocal microscopy and three-dimensional reconstruction of electrophysiologically identified neurons in thick brain slices. Journal of Electron Microscopy Technique, 18(1), 11–23.
Turner, J. N., Szarowski, D. H., Turner, T. J., Ancin, H., Lin, W. C., Roysam, B., et al. (1994). Three-dimensional imaging and image analysis of hippocampal neurons: confocal and digitally enhanced wide field microscopy. Microscopy Research and Technique, 29(4), 269–278.
Tyrrell, J. A., di Tomaso, E., Fuja, D., Tong, R., Kozak, K., Jain, R. K., et al. (2007). Robust 3-D modeling of vasculature imagery using superellipsoids. IEEE Transactions on Medical Imaging, 26(2), 223–237.
Tyrrell, J. A., Mahadevan, V., Tong, R. T., Brown, E. B., Jain, R. K., & Roysam, B. (2005). A 2-D/3-D model-based method to quantify the complexity of microvasculature imaged by in vivo multiphoton microscopy. Microvascular Research, 70(3), 165–178.
Vliet, L. J. V. (1993). Grey-scale measurements in multi-dimensional digitized images. Ph.D. Thesis of the Pattern Recognition Group, Delft University of Technology, The Netherlands.
Wearne, S. L., Rodriguez, A., Ehlenberger, D. B., Rocher, A. B., Henderson, S. C., & Hof, P. R. (2005). New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience, 136(3), 661–680.
Weaver, C. M., Hof, P. R., Wearne, S. L., & Lindquist, W. B. (2004). Automated algorithms for multiscale morphometry of neuronal dendrites. Neural Computation, 16(7), 1353–1383.
Wu, C. C., Reilly, J. F., Young, W. G., Morrison, J. H., & Bloom, F. E. (2004). High-throughput morphometric analysis of individual neurons. Cerebral Cortex, 14(5), 543–554.
Xiong, G., Zhou, X., Degterev, A., Ji, L., & Wong, S. T. (2006). Automated neurite labeling and analysis in fluorescence microscopy images. Cytometry A, 69(6), 494–505.
Xu, X., & Wong, S. T. (2006). Optical microscopic image processing of dendritic spines morphology. IEEE Signal Processing Magazine, 23(4), 132–135.
Yu, Z., & Bajaj, C. (2004). A segmentation-free approach for skeletonization of gray-scale images via anisotropic vector diffusion. Computer Vision and Pattern Recognition, CVPR. Proceedings of the 2004 IEEE Computer Society Conference on, 1, 415–20.
Yuste, R., & Bonhoeffer, T. (2001). Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annual Review of Neuroscience, 24, 1071–1089.
Zhang, Y., Zhou, X., Degterev, A., Lipinski, M., Adjeroh, D., Yuan, J., et al. (2007a). A novel tracing algorithm for high throughput imaging screening of neuron-based assays. Journal of Neuroscience Methods, 160(1), 149–162.
Zhang, Y., Zhou, X., Witt, R. M., Sabatini, B. L., Adjeroh, D., & Wong, S. T. (2007b). Automated spine detection using curvilinear structure detector and LDA Classifier. Biomedical Imaging: From Nano to Macro, ISBI. 4th IEEE International Symposium on, pp. 528–531.
Zhang, Y., Zhou, X., Witt, R. M., Sabatini, B. L., Adjeroh, D., & Wong, S. T. (2007c). Dendritic spine detection using curvilinear structure detector and LDA classifier. Neuroimage, 36(2), 346–360.
Acknowledgements
The image analysis aspects of this work were supported by NIH Biomedical Research Partnerships Grant R01 EB005157, by the Bernard M. Gordon Center for Subsurface Sensing and Imaging Systems, under the Engineering Research Centers Program of the National Science Foundation (Award Number EEC-9986821), and by Rensselaer Polytechnic Institute. The Potter laboratory images were collected by SMP and David Kantor in collaboration with Erin Schuman and Scott Fraser. Trachtenberg lab work was supported by NIMH grant P50 MH077972.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yuan, X., Trachtenberg, J.T., Potter, S.M. et al. MDL Constrained 3-D Grayscale Skeletonization Algorithm for Automated Extraction of Dendrites and Spines from Fluorescence Confocal Images. Neuroinform 7, 213–232 (2009). https://doi.org/10.1007/s12021-009-9057-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-009-9057-y