Abstract
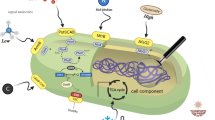
The drug discovery rate is dramatically decreasing due to the rediscovery of known compounds. Genome mining approaches have revealed that a large portion of the actinobacterial genome that encodes bioactive metabolites is cryptic and not expressed under standard lab conditions. In the present study, we aimed to induce antibiotic encoding biosynthetic genes in a member of Micrococcales as a new species of Promicromonospora, Promicromonospora kermanensis, by chemical and biological elicitors as it was considered to produce numerous valuable bioactive metabolites based on the whole genome results. Induction has been done via chemical (antibiotics, histone deacetylase inhibitors (HDAIs), rare earth elements (REEs), fatty acid synthesis inhibitors, and extreme pH changes) and biological elicitors (live and dead Gram-positive and negative bacteria). The results showed that valproic acid (as HDAIs), DMSO, lanthanum chloride (as REE), triclosan (as fatty acid synthesis inhibitors), alkaline pH, and supernatant of Pseudomonas aeruginosa UTMC 1404 culture could act as stimuli to provoke antibacterial synthetic pathways in Promicromonospora kermanensis DSM 45485. Moreover, it was revealed that eliciting agents in cell filtrated of P. aeruginosa culture is resistant to detergent, acidic, and basic condition and have amphipathic nature. The inducing effect of alkaline pH on metabolite induction of Actinobacteria was first reported in this study. In the follow-up studies, the induced antibacterial producing clusters can be subjected to the characterization, and the implemented approach in this study can be used for metabolites induction in other selected species.


Similar content being viewed by others
References
Zaman, S. B., Hussain, M. A., Nye, R., Mehta, V., Mamun, K. T., & Hossain, N. (2017). A review on antibiotic resistance: alarm bells are ringing. Cureus, 9(6), 1403.
O’Neil, J. (2014). Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Review on Antimicrobial Resistance, 20, 1–16.
Wink, J., Mohammadipanah, F., & Hamedi, J (2017). Biology and biotechnology of actinobacteria. Springer, Berlin.
Genilloud, O. (2019). Natural products discovery and potential for new antibiotics. Current Opinion in Microbiology, 51, 81–87.
Mohamed, A., Nguyen, C. H., & Mamitsuka, H. (2015). Current status and prospects of computational resources for natural product dereplication: a review. Briefings in Bioinformatics, 17(2), 309–321.
Seyedsayamdost, M. R. (2014). High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proceedings of the National Academy of Sciences, 111(20), 7266–7271.
Romano, S., Jackson, S. A., Patry, S., & Dobson, A. D. (2018). Extending the “one strain many compounds”(OSMAC) principle to marine microorganisms. Marine Drugs, 16(7), 244.
Baltz, R. H. (2016). Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. Journal of Industrial Microbiology & Biotechnology, 43(2-3), 343–370.
Hug, J. J., Bader, C. D., Remškar, M., Cirnski, K., & Müller, R. (2018). Concepts and methods to access novel antibiotics from actinomycetes. Antibiotics, 7(2), 44.
Bode, H. B., Bethe, B., Höfs, R., & Zeeck, A. (2002). Big effects from small changes: possible ways to explore nature's chemical diversity. ChemBioChem, 3(7), 619–627.
Covington, B. C., Spraggins, J. M., Ynigez-Gutierrez, A. E., Hylton, Z. B., & Bachmann, B. O. (2018). Response of hypogean actinobacterial genera secondary metabolism to chemical and biological stimuli. Applied and Environmental Microbiology, 84(19), e01125–18.
Tawfike, A., Attia, E. Z., Desoukey, S. Y., Hajjar, D., Makki, A. A., Schupp, P. J., Edrada-Ebel, R., & Abdelmohsen, U. R. (2019). New bioactive metabolites from the elicited marine sponge-derived bacterium Actinokineospora spheciospongiae sp. nov. AMB Express, 9(1), 12.
Van der Meij, A., Willemse, J., Schneijderberg, M. A., Geurts, R., Raaijmakers, J. M., & van Wezel, G. P. (2018). Inter-and intracellular colonization of Arabidopsis roots by endophytic actinobacteria and the impact of plant hormones on their antimicrobial activity. Antonie Van Leeuwenhoek, 111(5), 679–690.
Takano, E. (2006). γ-Butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Current Opinion in Microbiology, 9(3), 287–294.
Haferburg, G., & Kothe, E. (2013). Activation of silent genes in actinobacteria by exploiting metal stress, Actinobacteria. Application in bioremediation and production of industrial enzymes (pp. 56–73). Boca Raton: CRC Press.
Xu, D., Han, L., Li, C., Cao, Q., Zhu, D., Barrett, N. H., Harmody, D., Chen, J., Zhu, H., & McCarthy, P. J. (2018). Bioprospecting deep-sea actinobacteria for novel anti-infective natural products. Frontiers in Microbiology, 9, 787.
Kim, Y. J., Song, J. Y., Moon, M. H., Smith, C. P., Hong, S.-K., & Chang, Y. K. (2007). pH shock induces overexpression of regulatory and biosynthetic genes for actinorhodin production in Streptomyces coelicolor A3 (2). Applied Microbiology and Biotechnology, 76(5), 1119–1130.
Hassan, S. S. u., Jin, H.-Z., Abu-Izneid, T., Rauf, A., Ishaq, M., & Suleria, H. A. R. (2019). Stress-driven discovery in the natural products: A gateway towards new drugs. Biomedicine & Pharmacotherapy, 109, 459–467.
Moore, J. M., Bradshaw, E., Seipke, R. F., Hutchings, M. I., & McArthur, M. (2012). Use and discovery of chemical elicitors that stimulate biosynthetic gene clusters in Streptomyces bacteria. Methods in Enzymology. Elsevier, 517, 367–385.
Ochi, K., & Hosaka, T. (2013). New strategies for drug discovery: Activation of silent or weakly expressed microbial gene clusters. Applied Microbiology and Biotechnology, 97(1), 87–98.
So, A. G., & Davie, E. W. (1964). The effects of organic solvents on protein biosynthesis and their influence on the amino acid code. Biochemistry, 3(8), 1165–1169.
Chen, G., Li, X., Waters, B., & Davies, J. (2000). Enhanced production of microbial metabolites in the presence of dimethyl sulfoxide. The Journal of Antibiotics, 53(10), 1145–1153.
Zhu, L.-W., Zhong, J.-J., & Tang, Y.-J. (2008). Significance of fungal elicitors on the production of ganoderic acid and Ganoderma polysaccharides by the submerged culture of medicinal mushroom Ganoderma lucidum. Process Biochemistry, 43(12), 1359–1370.
Antoraz, S., Santamaría, R. I., Díaz, M., Sanz, D., & Rodríguez, H. (2015). Toward a new focus in antibiotic and drug discovery from the Streptomyces arsenal. Frontiers in Microbiology, 6, 461.
Sidda, J. D., & Corre, C. (2012). Gamma-butyrolactone and furan signaling systems in Streptomyces. Methods in Enzymology. Elsevier, 517, 71–87.
Garg, N., Manchanda, G., & Kumar, A. (2014). Bacterial quorum sensing: circuits and applications. Antonie Van Leeuwenhoek, 105(2), 289–305.
Tanaka, Y., Izawa, M., Hiraga, Y., Misaki, Y., Watanabe, T., & Ochi, K. (2017). Metabolic perturbation to enhance polyketide and nonribosomal peptide antibiotic production using triclosan and ribosome-targeting drugs. Applied Microbiology and Biotechnology, 101(11), 4417–4431.
Mohammadipanah, F., del Carmen Montero-Calasanz, M., Schumann, P., Spröer, C., Rohde, M., & Klenk, H.-P. (2017). Promicromonospora kermanensis sp. nov., an actinobacterium isolated from soil. International Journal of Systematic and Evolutionary Microbiology, 67(2), 262–267.
Okada, B. K., Wu, Y., Mao, D., Bushin, L. B., & Seyedsayamdost, M. R. (2016). Mapping the trimethoprim-induced secondary metabolome of Burkholderia thailandensis. ACS Chemical Biology, 11(8), 2124–2130.
Craney, A., Ozimok, C., Pimentel-Elardo, S. M., Capretta, A., & Nodwell, J. R. (2012). Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chemistry & Biology, 19(8), 1020–1027.
Olano, C., Lombo, F., Méndez, C., & Salas, J. A. (2008). Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metabolic Engineering, 10(5), 281–292.
Patterson, G. M., & Bolis, C. M. (1997). Fungal cell wall polysaccharides elicit an antifungal secondary metabolite (phytoalexin) in the cyanobacterium scytonema ocelutum 2. Journal of Phycology, 33(1), 54–60.
Fisch, K., Gillaspy, A., Gipson, M., Henrikson, J., Hoover, A., Jackson, L., Najar, F., Wägele, H., & Cichewicz, R. (2009). Chemical induction of silent biosynthetic pathway transcription in Aspergillus niger. Journal of Industrial Microbiology & Biotechnology, 36(9), 1199–1213.
Bok, J. W., Chiang, Y.-M., Szewczyk, E., Reyes-Dominguez, Y., Davidson, A. D., Sanchez, J. F., Lo, H.-C., Watanabe, K., Strauss, J., & Oakley, B. R. (2009). Chromatin-level regulation of biosynthetic gene clusters. Nature Chemical Biology, 5(7), 462–464.
Tyurin, A. P., Alferova, V. A., & Korshun, V. A. (2018). Chemical elicitors of antibiotic biosynthesis in actinomycetes. Microorganisms, 6(2), 52.
Song, L., Barona-Gomez, F., Corre, C., Xiang, L., Udwary, D. W., Austin, M. B., Noel, J. P., Moore, B. S., & Challis, G. L. (2006). Type III polyketide synthase β-ketoacyl-ACP starter unit and ethylmalonyl-CoA extender unit selectivity discovered by Streptomyces coelicolor genome mining. Journal of the American Chemical Society, 128(46), 0.14754–0.14755.
Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A.-M., Challis, G. L., Thomson, N., James, K. D., Harris, D. E., Quail, M. A., Kieser, H., & Harper, D. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2). Nature, 417(6885), 141–147.
Pettit, R. K. (2011). Small-molecule elicitation of microbial secondary metabolites. Microbial Biotechnology, 4(4), 471–478.
Kawai, K., Wang, G., Okamoto, S., & Ochi, K. (2007). The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiology Letters, 274(2), 311–315.
Tanaka, Y., Hosaka, T., & Ochi, K. (2010). Rare earth elements activate the secondary metabolite–biosynthetic gene clusters in Streptomyces coelicolor A3 (2). The Journal of Antibiotics, 63(8), 477–481.
Hayes, A., Hobbs, G., Smith, C. P., Oliver, S. G., & Butler, P. R. (1997). Environmental signals triggering methylenomycin production by Streptomyces coelicolor A3 (2). Journal of Bacteriology, 179(17), 5511–5515.
Horinouchi, S. (2003). AfsR as an integrator of signals that are sensed by multiple serine/threonine kinases in Streptomyces coelicolor A3 (2). Journal of Industrial Microbiology and Biotechnology, 30(8), 462–467.
Mohammadipanah, F, & Zamanzadeh, M. (2019). Bacterial mechanisms promoting the tolerance to drought stress in plants, In secondary metabolites of plant growth promoting Rhizomicroorganisms (Harikesh Bahadur Singh H. B., Keswani C., Reddy M. S., Sansinenea E. & García-Estrada C.), Springer, Singapore, pp.185–224.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 2231 kb)
Rights and permissions
About this article
Cite this article
Mohammadipanah, F., Kermani, F. & Salimi, F. Awakening the Secondary Metabolite Pathways of Promicromonospora kermanensis Using Physicochemical and Biological Elicitors. Appl Biochem Biotechnol 192, 1224–1237 (2020). https://doi.org/10.1007/s12010-020-03361-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03361-3