Abstract
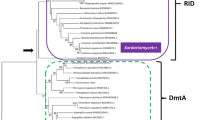
Manipulation of the fungal epigenome is hypothesized to be an effective method for accessing natural products from silent biosynthetic pathways. A library of epigenetic modifiers was tested using the fungus Aspergillus niger to determine the impact of small-molecule inhibitors on reversing the transcriptional suppression of biosynthetic genes involved in polyketide (PKS), non-ribosomal peptide (NRPS), and hybrid PKS-NRPS (HPN) production. Examination of expressed sequence tag libraries from A. niger demonstrated that >70% of its PKS-, NRPS-, and HPN-encoding gene clusters were transcriptionally suppressed under standard laboratory culture conditions. Using a chemical epigenetic methodology, we showed that treatment of A. niger with suberoylanilide hydroxamic acid and 5-azacytidine led to the transcriptional upregulation of many secondary-metabolite-encoding biosynthetic gene clusters. Chemical epigenetic modifiers exhibited positional biases for upregulating chromosomally distal gene clusters. In addition, a phylogenetic-based preference was noted in the upregulation of reducing clade I PKS gene clusters, while reducing clade IV PKS gene clusters were largely unaffected. Manipulating epigenetic features in fungi is a powerful method for accessing the products of silent biosynthetic pathways. Moreover, this approach can be readily incorporated into modern microbial screening operations.





Similar content being viewed by others
References
Wilkinson B, Micklefield J (2007) Mining and engineering natural-product biosynthetic pathways. Nat Chem Biol 3:379–386. doi:10.1038/nchembio.2007.7
Scherlach K, Hertweck C (2009) Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem 7:1753–1760. doi:10.1039/b821578b
Zerikly M, Challis GL (2009) Strategies for the discovery of new natural products by genome mining. Chem Bio Chem 10:625–633. doi:10.1002/cbic.200800389
Kamper J, Kahmann R, Bolker M, Ma L-J, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Muller O, Perlin MH, Wosten HAB, de Vries R, Ruiz-Herrera J, Reynaga-Pena CG, Snetselaar K, McCann M, Perez-Martin J, Feldbrugge M, Basse CW, Steinberg G, Ibeas JI, Holloman W, Guzman P, Farman M, Stajich JE, Sentandreu R, Gonzalez-Prieto JM, Kennell JC, Molina L, Schirawski J, Mendoza-Mendoza A, Greilinger D, Munch K, Rossel N, Scherer M, Vranes M, Ladendorf O, Vincon V, Fuchs U, Sandrock B, Meng S, Ho ECH, Cahill MJ, Boyce KJ, Klose J, Klosterman SJ, Deelstra HJ, Ortiz-Castellanos L, Li W, Sanchez-Alonso P, Schreier PH, Hauser-Hahn I, Vaupel M, Koopmann E, Friedrich G, Voss H, Schluter T, Margolis J, Platt D, Swimmer C, Gnirke A, Chen F, Vysotskaia V, Mannhaupt G, Guldener U, Munsterkotter M, Haase D, Oesterheld M, Mewes H-W, Mauceli EW, DeCaprio D, Wade CM, Butler J, Young S, Jaffe DB, Calvo S, Nusbaum C, Galagan J, Birren BW (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444:97–101. doi:10.1038/nature05248
Bölker M, Basse CW, Schirawski J (2008) Ustilago maydis secondary metabolism—from genomics to biochemistry. Fungal Gen Biol 45:S88–S93. doi:10.1016/j.fgb.2008.05.007
Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma L-J, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CPC, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, Birren B (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859–868. doi:10.1038/nature01554
Galagan JE, Calvo SE, Cuomo C, Ma L-J, Wortman JR, Batzoglou S, Lee S-I, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW (2005) Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115. doi:10.1038/nature04341
Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N (2005) Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156. doi:10.1038/nature04332
Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JAE, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EGJ, Debets AJM, Dekker P, van Dijck PWM, van Dijk A, Dijkhuizen L, Driessen AJM, d’Enfert C, Geysens S, Goosen C, Groot GSP, de Groot PWJ, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JPTW, van den Hondel CAMJJ, van der Heijden RTJM, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJEC, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NNME, Ram AFJ, Rinas U, Roubos JA, Sagt CMJ, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJJ, Wedler H, Wosten HAB, Zeng A-P, van Ooyen AJJ, Visser J, Stam H (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25:221–231. doi:10.1038/nbt1282
Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu J-R, Pan H, Read ND, Lee Y-H, Carbone I, Brown D, Oh YY, Donofrio N, Jeong JS, Soanes DM, Djonovic S, Kolomiets E, Rehmeyer C, Li W, Harding M, Kim S, Lebrun M-H, Bohnert H, Coughlan S, Butler J, Calvo S, Ma L-J, Nicol R, Purcell S, Nusbaum C, Galagan JE, Birren BW (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980–986. doi:10.1038/nature03449
Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K-I, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J-I, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161. doi:10.1038/nature04300
Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH (2008) Epigenetic remodeling of the fungal secondary metabolome. Org Biomol Chem 6:1895–1897. doi:10.1039/b804701d
Henrikson JC, Hoover AR, Joyner PM, Cichewicz RH (2009) A chemical epigenetics approach for engineering the in situ biosynthesis of a cryptic natural product from Aspergillus niger. Org Biomol Chem 7:435–438. doi:10.1039/b819208a
Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP (2007) Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell 6:1656–1664. doi:10.1128/ec.00186-07
Grewal SIS, Bonaduce MJ, Klar AJS (1998) Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150:563–576
Bernstein BE, Tong JK, Schreiber SL (2000) Genomewide studies of histone deacetylase function in yeast. Proc Nat Acad Sci USA 97:13708–13713. doi:10.1073/pnas.250477697
Graessle S, Dangl M, Haas H, Mair K, Trojer P, Brandtner E-M, Walton JD, Loidl P, Brosch G (2000) Characterization of two putative histone deacetylase genes from Aspergillus nidulans. Biochim Biophys Acta 1492:120–126. doi:10.1016/S0167-4781(00)00093-2
Peng W, Togawa C, Zhang K, Kurdistani SK (2008) Regulators of cellular levels of histone acetylation in Saccharomyces cerevisiae. Genetics 179:277–289. doi:10.1534/genetics.107.085068
Tamaru H, Selker EU (2001) A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277–283. doi:10.1038/35104508
Trojer P, Brandtner EM, Brosch G, Loidl P, Galehr J, Linzmaier R, Haas H, Mair K, Tribus M, Graessle S (2003) Histone deacetylases in fungi: novel members, new facts. Nucleic Acids Res 31:3971–3981
van West P, Shepherd SJ, Walker CA, Li S, Appiah AA, Grenville-Briggs LJ, Govers F, Gow NAR (2008) Internuclear gene silencing in Phytophthora infestans is established through chromatin remodelling. Microbiol 154:1482–1490. doi:10.1099/mic.0.2007/015545-0
Brosch G, Loidl P, Graessle S (2008) Histone modifications and chromatin dynamics: a focus on filamentous fungi. FEMS Microbiol Rev 32:409–439. doi:10.1111/j.1574-6976.2007.00100.x
Biel M, Wascholowski V, Giannis A (2005) Epigenetics—an epicenter of gene regulation: histones and histone-modifying enzymes. Angew Chem Int Ed 44:3186–3216. doi:10.1002/anie.200461346
Cole PA (2008) Chemical probes for histone-modifying enzymes. Nat Chem Biol 4:590–597. doi:10.1038/nchembio.111
Goffin J, Eisenhauer E (2002) DNA methyltransferase inhibitors—state of the art. Ann Oncol 2002(13):1699–1716
Zhou Y, Cambareri EB, Kinsey JA (2001) DNA methylation inhibits expression and transposition of the Neurospora Tad retrotransposon. Mol Genet Genomics 265:748–754. doi:10.1007/s004380100472
Lee DW, Freitag M, Selker EU, Aramayo R (2008) A cytosine methyltransferase homologue is essential for sexual development in Aspergillus nidulans. PLoS ONE 3:e2531. doi:10.1371/journal.pone.0002531
Wada H, Kagoshima M, Ito K, Barnes PJ, Adcock IM (2005) 5-Azacytidine suppresses RNA polymerase II recruitment to the SLPI gene. Biochem Biophys Res Comm 331:93–99. doi:10.1016/j.bbrc.2005.03.138
Wozniak RJ, Klimecki WT, Lau SS, Feinstein Y, Futscher BW (2006) 5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene 26:77–90. doi:10.1038/sj.onc.1209763
Andersen MR, Nielsen J (2009) Current status of systems biology in Aspergilli. Fungal Genet Biol 46:S180–S190. doi:10.1016/j.fgb.2008.07.006
Baker SE (2006) Aspergillus niger genomics: past, present and into the future. Med Mycol 44:17–21
Primer3 (v. 0.4.0) Available at: http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 25:402–408. doi:10.1006/meth.2001.1262
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Semova N, Storms R, John T, Gaudet P, Ulycznyj P, Min X, Sun J, Butler G, Tsang A (2006) Generation, annotation, and analysis of an extensive Aspergillus niger EST collection. BMC Microbiol 6:7. doi:10.1186/1471-2180-6-7
Bohle K, Jungebloud A, Göcke Y, Dalpiaz A, Cordes C, Horn H, Hempel DC (2007) Selection of reference genes for normalisation of specific gene quantification data of Aspergillus niger. J Biotechnol 132:353–358. doi:10.1016/j.jbiotec.2007.08.005
Van Guilder HD, Vrana KE, Freeman WM (2008) Twenty-five years of quantitative PCR for gene expression analysis. BioTechniques 44:619–626
Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, Parkhill J (2005) ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423. doi:10.1093/bioinformatics/bti553
Cox RJ (2007) Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem 5:2010–2026. doi:10.1039/b704420h
Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG (2003) Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Nat Acad Sci USA 100:15670–15675. doi:10.1073/pnas.2532165100
Amnuaykanjanasin A, Punya J, Paungmoung P, Rungrod A, Tachaleat A, Pongpattanakitshote S, Cheevadhanarak S, Tanticharoen M (2005) Diversity of type I polyketide synthase genes in the wood-decay fungus Xylaria sp. BCC 1067. FEMS Microbiol Lett 251:125–136. doi:10.1016/j.femsle.2005.07.038
Crawford JM, Vagstad AL, Ehrlich KC, Townsend CA (2008) Starter unit specificity directs genome mining of polyketide synthase pathways in fungi. Bioorg Chem 36:16–22. doi:10.1016/j.bioorg.2007.11.002
James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung G-H, Johnson D, O/’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schuszler A, Longcore JE, O/’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lucking R, Budel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R (2006) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443:818–822. doi:10.1038/nature05110
Gross H (2007) Strategies to unravel the function of orphan biosynthesis pathways: recent examples and future prospects. Appl Microbiol Biotechnol 75:267–277. doi:10.1007/s00253-007-0900-5
Bok JW, Noordermeer D, Kale SP, Keller NP (2006) Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol Microbiol 61:1636–1645. doi:10.1111/j.1365-2958.2006.05330.x
Perrin RM, Fedorova ND, Bok JW, Cramer RA, Wortman JR, Kim HS, Nierman WC, Keller NP (2007) Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog 3:508–517. doi:10.1371/journal.ppat.0030050
Katan-Khaykovich Y, Struhl K (2005) Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J 24:2138–2149. doi:10.1038/sj.emboj.7600692
Lam KS (2007) New aspects of natural products in drug discovery. Trends Microbiol 15:279–289. doi:10.1016/j.tim.2007.04.001
Koehn FE, Carter GT (2005) The evolving role of natural products in drug discovery. Nat Rev Drug Disc 4:206–220. doi:10.1038/nrd1657
Paterson I, Anderson EA (2005) The renaissance of natural products as drug candidates. Science 310:451–453. doi:10.1126/science.1116364
Baker DD, Chu M, Oza U, Rajgarhia V (2007) The value of natural products to future pharmaceutical discovery. Nat Prod Rep 24:1225–1244. doi:10.1039/b602241n
Newman DJ, Cragg GM (2007) Natural products as sources of new drugs over the last 25 years. J Nat Prod 70:461–477. doi:10.1021/np068054v
Acknowledgments
This work was supported by the University of Oklahoma, College of Arts and Sciences and the University of Oklahoma Department of Chemistry and Biochemistry. We thank D. Dyer, J. Piel, and B. A. Roe for helpful comments throughout the preparation of this manuscript. We are also grateful for editorial and technical input from F. Schmitz during the writing of this paper.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fisch, K.M., Gillaspy, A.F., Gipson, M. et al. Chemical induction of silent biosynthetic pathway transcription in Aspergillus niger . J Ind Microbiol Biotechnol 36, 1199–1213 (2009). https://doi.org/10.1007/s10295-009-0601-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-009-0601-4