Abstract
Purpose of Review
The functions, genetic variations and impact of apolipoprotein E on lipoprotein metabolism in general are placed in the context of clinical practice dealing with moderate dyslipidaemia as well as dysbetalipoproteinemia, a highly atherogenic disorder and lipoprotein glomerulopathy.
Recent Findings
Additional variants of apolipoprotein E and participation of apolipoprotein E in inflammation are of interest. The mostly favourable effects of apolipoprotein E2 as well as the atherogenic nature of apolipoproteinE4, which has an association with cognitive impairment, are confirmed. The contribution of remnant lipoproteins of triglyceride-rich lipoproteins, of which dysbetalipoproteinemia represents an extreme, is explored in atherosclerosis. Mimetic peptides may present new therapeutic approaches.
Summary
Apolipoprotein E is an important determinant of the lipid profile and cardiovascular health in the population at large and can precipitate dysbetalipoproteinemia and glomerulopathy. Awareness of apolipoprotein E polymorphisms should improve medical care.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Mahley RW. Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders. J Mol Med. 2016;94:739–46. https://doi.org/10.1007/s00109-016-1427-yAn excellent historical and scientific account.
Marais AD. Apolipoprotein E in lipoprotein metabolism, health and cardiovascular disease. Pathology. 2019;51((2):165–76. https://doi.org/10.1016/j.pathol.2018.11.002Review of apoE variants on dyslipidemia.
Sniderman AD, Type III. Hyperlipoproteinemia: the forgotten, disregarded, neglected, overlooked, ignored but highly atherogenic, and highly treatable dyslipoproteinemia. Clin Chem. 2019;65(2):225–7. https://doi.org/10.1373/clinchem.2018.298026An important reminder of dysbetalipoproteinemia.
Bennett BJ, Davis RC, Civelek M, Orozco L, Wu J, Qi H, et al. Genetic architecture of atherosclerosis in mice: a systems genetics analysis of common inbred strains. PLoS Genet. 2015;11(12):e1005711. https://doi.org/10.1371/journal.pgen.1005711.
Getz GS, Reardon CA. ApoE knockout and knockin mice: the history of their contribution to the understanding of atherogenesis. J Lipid Res. 2016;57:758–66. https://doi.org/10.1194/jlr.R067249.
Huang L, Hua Z, Xiao H, Cheng Y, Xu K, Gao Q, et al. CRISPR/Cas9-mediated apoE-/- and LDLR-/- double gene knockout in pigs elevates serum LDL-C and TC levels. Oncotarget. 2017;8(23):37751–60. https://doi.org/10.18632/oncotarget.17154.
Shim J, Poulsen CB, Hagensen MK, Larsen T, Heegaard PMH, Christoffersen C, et al. Apolipoprotein E deficiency increases remnant lipoproteins and accelerates progressive atherosclerosis, but not xanthoma formation, in gene-modified minipigs. JACC Basic Transl Sci. 2017;2(5):591–600. https://doi.org/10.1016/j.jacbts.2017.06.004.
Wagner T, Bartelt A, Schlein C, Heeren J. Genetic dissection of tissue-specific apolipoprotein E function for hypercholesterolemia and diet-induced obesity. PLoS One. 2015:1–14. https://doi.org/10.1371/journal.pone.0145102A demonstration of the importance of apoE in the liver.
Tudorache IF, Trusca VG, Gafencu AV. Apolipoprotein E—a multifunctional protein with implications in various pathologies as a result of its structural features. Comput Struct Biotechnol J. 2017;1:359–65. https://doi.org/10.1016/j.csbj.2017.05.003.
Benevides LD, de Carvalho DS, Andrade RFS, Bomfim GC, de Campos Fernandes FM. Evolutionary analysis of apolipoprotein E by Maximum Likelihood and complex network methods. Genet Mol Biol. 2016;39(4):665–73. https://doi.org/10.1590/1678-4685-GMB-2015-016.
Takacs CN, Andreo U, Belote RL, Pulupa J, Scull MA, Gleason CE, et al. GFP-tagged apolipoprotein E: a useful marker for the study of hepatic lipoprotein egress. Traffic. 2017;18(3):192–204. https://doi.org/10.1111/tra.12467.
Chetty PS. Leland Mayneb, Lund-Katz S, Englander SW, Phillip MC. Helical structure, stability, and dynamics in human apolipoprotein E3 and E4 by hydrogen exchange and mass spectrometry. Proc Natl Acad Sci U S A. 2017;114(5):968–73. https://doi.org/10.1073/pnas.1617523114Full description of apoE self-association and formation of HDL.
Hubin E, Verghese PB, van Nuland N, Broersen K. Apolipoprotein E associated with reconstituted high-density lipoprotein-like particles is protected from aggregation. FEBS Lett. 2019;593:1144–53. https://doi.org/10.1002/1873-3468.
Lanfranco MF, Ng CA, Rebeck GW. ApoE lipidation as a therapeutic target in Alzheimer’s disease. Int J Mol Sci. 2020;21:6336. https://doi.org/10.3390/ijms21176336Informative about apoE in lipid homoestasis in neurons.
Melhem H, Kallol S, Huang X, Lüthi M, Ontsouka CE, Keogh A, et al. Placental secretion of apolipoprotein A1 and E: the antiatherogenic impact of the placenta. Sci Rep. 2019;9:6225. https://doi.org/10.1038/s41598-019-42522.
Norda S, Rausch TK, Orlikowsky T, Hütten M, Schulz S, Göpel W, et al. Apolipoprotein E genotype in very preterm neonates with intrauterine growth restriction: an analysis of the german neonatal network cohort. Biomed Res Int. 2017;2017:2837027, 8 pages. https://doi.org/10.1155/2017/2837027.
Mak ACY, Pullinger CR, Tang LF, Wong JS, Deo RC, Schwarz J-M, et al. Effects of the absence of apolipoprotein E on lipoproteins, neurocognitive function, and retinal function. JAMA Neurol. 2014;71(10):1228–36. https://doi.org/10.1001/jamaneurol.2014.2011.
Pirim D, Radwan ZH, Wang X, Niemsiri V, Hokanson JE, Hamman RF, et al. Apolipoprotein E-C1-C4-C2 gene cluster region and inter-individual variation in plasma lipoprotein levels: a comprehensive genetic association study in two ethnic groups. PLoS One. 14(3):e0214060. https://doi.org/10.1371/journal.pone.0214060The description of common variants affecting apoE.
Low-Kam C, Rhainds D, Lo KS, Barhdadi A, Boul M, Alem S, et al. Variants at the APOE/C1/C2/C4 locus modulate cholesterol efflux capacity independently of high-density lipoprotein cholesterol. J Am Heart Assoc. 2018;7:e009545. https://doi.org/10.1161/JAHA.118.009545.
Theendakara V, Peters-Libeu CA, Spilman P, Poksay KS, Bredesen DE, Rao RV. Direct transcriptional effects of apolipoprotein E. J Neurosci. 2016;36(3):685–700. https://doi.org/10.1523/JNEUROSCI.3562-15.2016.
Babenko VN, Afonnikov DA, Ignatieva EV, Klimov AV, Gusev FE, Rogaev EI. Haplotype analysis of APOE intragenic SNPs. BMC Neurosci. 2018;19(Suppl 1):16. https://doi.org/10.1186/s12868-018-0413-4A very detailed description of variation apoE gene in populations.
Trusca VG, Fuior EV, Kardassis D, Simionescu M, Gafencu AV. The opposite effect of c-Jun transcription factor on apolipoprotein E gene regulation in hepatocytes and macrophages. Int J Mol Sci. 2019;20:1471. https://doi.org/10.3390/ijms20061471.
Benson MD, Yang Q, Ngo D, Zhu Y, Shen D, Farrell LA, et al. The genetic architecture of the cardiovascular risk proteome. Circulation. 2018;137(11):1158–72. https://doi.org/10.1161/CIRCULATIONAHA.117.029536.
Mancera-Páez O, Estrada-Orozco K, Mahecha MF, Cruz F, Bonilla-Vargas K, Sandoval N, et al. Differential methylation in APOE (Chr19; Exon Four; from 44,909,188 to 44,909,373/hg38) and increased apolipoprotein E plasma levels in subjects with mild cognitive impairment. Int J Mol Sci. 2019;20(6):1394. Published online 2019 Mar 20. https://doi.org/10.3390/ijms20061394.
Ma Y, Smith CE, Lai C-Q, Irvin MR, Parnell LD, Lee Y-C, et al. Genetic variants modify the effect of age on APOE methylation in the Genetics of Lipid Lowering Drugs and Diet Network study. Aging Cell. 2015;14:49–59. https://doi.org/10.1111/acel.12293.
Karlsson IK, Ploner A, Wang Y, Gatz M, Pedersen NL, Hägg S. Apolipoprotein E DNA methylation and late-life disease. Int J Epidemiol. 2018;47:899–907. https://doi.org/10.1093/ije/dyy025.
Mur J, McCartney DL, Walker RM, Campbell A, Bermingham ML, Morris SW, et al. DNA methylation in APOE: the relationship with Alzheimer’s and with cardiovascular health. Alzheimers Dement. 2020;6:e12026. https://doi.org/10.1002/trc2.12026.
Rebeck GW. The role of APOE on lipid homeostasis and inflammation in normal brains. J Lipid Res. 2017;58:1493–9. https://doi.org/10.1194/jlr.R075408Multiple aspects of lipid metabolism and apoE including post-translational modification. Broadly informative of apoE and particularly on inflammation and discusses lower levels of apoE4.
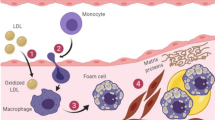
Getz GS, Reardon CA. Apoproteins E, A-I, and SAA in Macrophage pathobiology related to atherogenesis. Front Pharmacol. 2019;10:536. https://doi.org/10.3389/fphar.2019.00536An in-depth review of inflammation relevant to apoE and macrophages.
Centa M, Prokopec KE, Garimella MG, Habir K, Hofste L, Stark JM, et al. Acute loss of apolipoprotein E triggers an autoimmune response that accelerates atherosclerosis. Arterioscler Thromb Vasc Biol. 2018;38(8):e145–58. https://doi.org/10.1161/ATVBAHA.118.310802.
Giordano-Mooga S, Datta G, Wolkowicz P, Garber DW, Palgunachari M, White CR, et al. Apolipoprotein E mimetic peptide AEM-2 attenuates mitochondrial injury and apoptosis in human THP-1 macrophages. Curr Top Pept Protein Res. 2018;19:15–25.
Yin C, Ackermann S, Ma Z, Mohanta SK, Zhang C, Li Y, et al. ApoE attenuates unresolvable inflammation by complex formation with activated C1q. Nat Med. 2019;25(3):496–506. https://doi.org/10.1038/s41591-018-0336-8.
Nissilä E, Hakala P, Leskinen K, Roig A, Syed S, Van Kessel KPM, et al. Complement factor H and apolipoprotein E participate in regulation of inflammation in THP-1 macrophages. Front Immunol. 2018;9:2701. https://doi.org/10.3389/fimmu.2018.02701.
Bouchareychas L, Raffai RL. apolipoprotein E and atherosclerosis: from lipoprotein metabolism to microRNA control of inflammation. J Cardiovasc Dev Dis. 2018;5:30. https://doi.org/10.3390/jcdd5020030A useful review including pleiotropic effects of apoE.
Teter B, Ladu MJ, Sullivan PM, Frautschy SA, Cole GM. Apolipoprotein E isotype-dependent modulation of microRNA-146a in plasma and brain. Neuroreport. 2016;27(11):791–5. https://doi.org/10.1097/WNR.0000000000000608.
Zhong H, Cai Y, Cheng J, Cai D, Chen L, Su C, et al. Apolipoprotein E epsilon 4 enhances the association between the rs2910164 polymorphism of miR-146a and risk of atherosclerotic cerebral infarction. J Atheroscler Thromb. 2016;23:819–29. https://doi.org/10.5551/jat.32904.
Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9:19. https://doi.org/10.1186/s13578-019-0282-2.
Peng KY, Perez-Gonzalez R, Alldred MJ, Goulbourne CN, Morales-Corraliza J, Saito M, et al. Apolipoprotein E4 genotype compromises brain exosome production. Brain. 2019;142:163–75. https://doi.org/10.1093/brain/awy289.
Sack FM. The crucial roles of apolipoproteins E and C-III in apoB lipoprotein metabolism in normolipidemia and hypertriglyceridemia. Curr Opin Lipidol. 2015;26(1):56–63. https://doi.org/10.1097/MOL.0000000000000146.
Ramms B, Gordts PLSM. Apolipoprotein C-III in triglyceriderich lipoprotein metabolism. Curr Opin Lipidol. 2018;29:171–9. https://doi.org/10.1097/MOL.0000000000000502.
TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371:22–31. https://doi.org/10.1056/NEJMoa1307095.
Jepsen AM, Langsted A, Varbo A, Bang LE, Kamstrup PR, Nordestgaard BG. Increased remnant cholesterol explains part of residual risk of all-cause mortality in 5414 patients with ischemic heart disease. Clin Chem. 2016;62(4):593–604. https://doi.org/10.1373/clinchem.2015.253757.
Lawler PR, Akinkuolie AO, Chu AY, Shah SH, Kraus WE, Craig D, et al. Atherogenic lipoprotein determinants of cardiovascular disease and residual risk among individuals with low low-density lipoprotein cholesterol. J Am Heart Assoc. 2017;6(7):e005549. https://doi.org/10.1161/JAHA.117.005549.
Tada H, Nohara A, Inazu A, Mabuchi H, Kawashiri M. Remnant lipoproteins and atherosclerotic cardiovascular disease. Clin Chim Acta. 2019;490:1–5. https://doi.org/10.1016/j.cca.2018.12.014A comprehensive review.
Lane-Donovan C, Herz J. ApoE, ApoE receptors, and the synapse in Alzheimer’s disease. Trends Endocrinol Metab. 2017;28(4):273–84. https://doi.org/10.1016/j.tem.2016.12.001Complexities of apoE metabolism in the nervous will also indicate potential effects elsewhere.
Fischer AW, Albers K, Krott LM, Hoffzimmer B, Heine M, Schmale H, et al. The adaptor protein PID1 regulates receptor dependent endocytosis of postprandial triglyceride-rich lipoproteins. Mol Metab. 2018;16:88e99. https://doi.org/10.1016/j.molmet.2018.07.010.
Anower-E-Khuda F, Singh G, Deng Y, Gordts PLSM, Esko JD. Triglyceride-rich lipoprotein binding and uptake by heparan sulfate proteoglycan receptors in a CRISPR/Cas9 library of Hep3B mutants. Glycobiology. 2019;29(8):582–92. https://doi.org/10.1093/glycob/cwz037.
Love-Gregory L, Kraja AT, Allum F, Aslibekyan S, Hedman AK, Duan Y, et al. Higher chylomicron remnants and LDL particle numbers associate with CD36 SNPs and DNA methylation sites that reduce CD36. J Lipid Res. 2016;57:2176–84. https://doi.org/10.1194/jlr.P065250.
Plochberger B, Sych T, Weber F, Novacek J, Axmann M, Stangl H, et al. Lipoprotein particles interact with membranes and transfer their cargo without receptors. Biochemistry. 2020;59:4421–8. https://doi.org/10.1021/acs.biochem.0c00748.
Heeren J, Beisiegel U, Grewal T. Apolipoprotein E recycling: Implications for dyslipidemia and atherosclerosis. Arterioscler Thromb Vasc Biol. 2006;26:442–8. https://doi.org/10.1161/01.ATV.0000201282.64751.47.
Yang H, Zhang N, Okoro EU, Guo Z. Transport of apolipoprotein B-containing lipoproteins through endothelial cells is associated with apolipoprotein E-carrying HDL-like particle formation. Int J Mol Sci. 2018;19:3593. https://doi.org/10.3390/ijms19113593.
Balling M, Langsted A, Afzal S, Varbo A, Smith GD, Nordestgaard BG. A third of nonfasting plasma cholesterol is in remnant lipoproteins: Lipoprotein subclass profiling in 9293 individuals. Atherosclerosis. 2019;286:97–104. https://doi.org/10.1016/j.atherosclerosis.2019.05.011A very complete analysis of lipids and lipoproteins in a large cohort.
Remaley AT, Otvos JD. Methodological issues regarding: “A third of nonfasting plasma cholesterol is in remnant lipoproteins: lipoprotein subclass profiling in 9293 individuals”. Atherosclerosis. 2020. https://doi.org/10.1016/j.atherosclerosis.2020.01.020.
Chen J, Kuang J, Tang X, Mao L, Guo X, Luo Q, et al. Comparison of calculated remnant lipoprotein cholesterol levels with levels directly measured by nuclear magnetic resonance. Lipids Health Dis. 2020;132:1–13. https://doi.org/10.1186/s12944-020-01311-w.
Hirao Y, Nakajima K, Machida T, Murakami M, Ito Y. Development of a novel homogenous assay for remnant lipoprotein particle cholesterol. J Appl Lab Med. 2018;3(1):26–36. https://doi.org/10.1373/jalm.2017.024919.
Saeed A, Feofanova EV, Yu B, Sun W, Virani SS, Nambi V, et al. Remnant-like particle cholesterol, low-density lipoprotein triglycerides and incident cardiovascular disease. J Am Coll Cardiol. 2018;72(2):156–69. https://doi.org/10.1016/j.jacc.2018.04.050.
Masuda D, Yamashita S. Postprandial hyperlipidemia and remnant lipoproteins. J Atheroscler Thromb. 2017;24:95–109. https://doi.org/10.5551/jat.RV16003.
Bagdade JD, Jilma B, Hudgins LC, Alaupovic P, McCurdy CE. LpA-II:B:C:D:E: a new immunochemically defined acute phase lipoprotein in humans. Lipids Health Dis. 2018;17:127. https://doi.org/10.1186/s12944-018-0769-6.
Ben-Aicha S, Badimon L, Vilahur G. Advances in HDL: much more than lipid transporters. Int J Mol Sci. 2020;21:732. https://doi.org/10.3390/ijms21030732An up-to-date review.
Kajani S, Curley S, McGillicuddy FC. Unravelling HDL—looking beyond the cholesterol surface to the quality within Int. J Mol Sci. 2018;19:1971. https://doi.org/10.3390/ijms190719711-23.
Singh SA, Andraski AB, Pieper B, Goh W, Mendivil CO, Sacks FM, et al. Multiple apolipoprotein kinetics measured in human HDL by high-resolution/accurate mass parallel reaction monitoring. J Lipid Res. 2016;57:714–28. https://doi.org/10.1194/jlr.D061432.
Trieb M, Horvath A, Birner-Gruenberger R, Spindelboeck W, Stadlbauer V, Taschler U, et al. Liver disease alters high-density lipoprotein composition, metabolism and function. Biochim Biophys Acta. 2016;1861(7):630–8. https://doi.org/10.1016/j.bbalip.
Rosales C, Tang D, Gillard BK, Courtney HS, Pownall HJ. Apolipoprotein E mediates enhanced plasma HDL cholesterol clearance by low dose streptococcal serum opacity factor via hepatic LDL receptors in vivo. Arterioscler Thromb Vasc Biol. 2011;31(8):1834–41. https://doi.org/10.1161/ATVBAHA.111.224360.
Moriarty PM, Varvel SA, Gords PLSM, McConnell JP, Tsimikas S. Lp(a) mass levels increase significantly according to APOE genotype: an analysis of 431,239 patients. Arterioscler Thromb Vasc Biol. 2017;37(3):580–8. https://doi.org/10.1161/ATVBAHA.116.308704.
Blanchard V, Croyal M, Khantalin I, Ramin-Mangata S, Chemello K, Nativel B, et al. Reduced lipoprotein(a) associated with the apolipoprotein E2 genotype confers cardiovascular protection in familial hypercholesterolemia. JACC Basic Transl Sci. 2019;4(3):425–7.
Hopkins PN, Brinton EA, Nanjee MN. Hyperlipoproteinemia type 3: the forgotten phenotype. Curr Atheroscler Rep. 2014;6:440. 1-12. https://doi.org/10.1007/s11883-014-0440-2.
Hazard WR, Porte D, Bierman EL. Abnormal lipid composition of very low density lipoproteins in diagnosis of broad-beta disease (type III hyperlipoproteinemia). Metabolism. 1972;21(11):1009–19. https://doi.org/10.1016/0026-0495(72)90031-5.
Blum CB. Type III hyperlipoproteinemia: still worth considering? Prog Cardiovasc Dis. 2016;59:119–24. https://doi.org/10.1016/j.pcad.2016.07.007.
Salinas CAA, Chapman MJ. Remnant lipoproteins: are they equal to or more atherogenic than LDL? Curr Opin Lipidol. 2020;31:132–9. https://doi.org/10.1097/MOL.0000000000000682.
Le R, Abbas M, McIntyre AD, Hegele RA. Severe combined dyslipidemia with a complex genetic basis. J Investig Med High Impact Case Rep. 2019;7:1–5. https://doi.org/10.1177/2324709619877050.
Sniderman AD, de Graaf J, Thanassoulis G, Tremblay AJ, Martin SS, Couture P. The spectrum of type III hyperlipoproteinemia. J Clin Lipidol. 2018;12:1383–9 Dysbetalipoproteinemia review and guides that apoB can discriminate dysbetalipoproteinemia.
Boot CS, Middling E, Allen J, Neely RDG. Evaluation of non-HDL cholesterol to apolipoprotein B ratio as a screening test for dysbetalipoproteinemia. Clin Chem. 2019;65:313–20. https://doi.org/10.1373/clinchem.2018.292425A method for diagnosis of dysbetalipoproteinemia from the conventional lipid profile.
Pallazola VA, Sathiyakumar V, Park J, Vakil RM, Toth PP, Lazo-Elizondo M, et al. Modern prevalence of dysbetalipoproteinemia (Fredrickson-Levy-Lees type III hyperlipoproteinemia). Arch Med Sci. 2020;16(5):993–1003. https://doi.org/10.5114/aoms.2019.86972.
Wintjens R, Bozon D, Belabbas K, Mbou F, Girardet J-P, Tounian P, et al. Global molecular analysis and APOE mutations in a cohort of autosomal dominant hypercholesterolemia patients in France. J Lipid Res. 2016;57:482–91. https://doi.org/10.1194/jlr.P055699.
Calandra S, Tarugi P, Bertolini S. Impact of rare variants in autosomal dominant hypercholesterolemia causing genes. Curr Opin Lipidol. 2017;28:267–72. https://doi.org/10.1097/MOL.0000000000000414.
Leren TP, Strøm TB, Berge KE. Variable phenotypic expression of nonsense mutation p.Thr5* in the APOE gene. Mol Genet Metab Rep. 2016;9:67–70. https://doi.org/10.1016/j.ymgmr.2016.10.007.
Kato T, Ushiogi Y, Yokoyama H, Hara S, Matsunaga A, Muso E, et al. A case of apolipoprotein E Toyonaka and homozygous apolipoprotein E2/2 showing non-immune membranous nephropathy-like glomerular lesions with foamy changes. CEN Case Rep. 2019;8:106–11. https://doi.org/10.1007/s13730-019-00380-w.
Wu H, Yang Y, Hu Z. The novel apolipoprotein E mutation apoE Chengdu (c.518T C, p.L173P) in a Chinese patient with lipoprotein glomerulopathy. J Atheroscler Thromb. 2018;25:733–40. https://doi.org/10.5551/jat.41996.
Sasaki M, Yasuno T, Ito K, Matsunaga A, Hisano S, Abe Y, et al. Focal segmental glomerulosclerosis with heterozygous apolipoprotein E5 (Glu3Lys). CEN Case Rep. 2018;7:225–8. https://doi.org/10.1007/s13730-018-0331-4.
Tavori H, Fan D, Giunzioni I, Zhu L, Linton MF, Fogo AB, et al. Macrophage-derived apoE Sendai suppresses atherosclerosis while causing lipoprotein glomerulopathy in hyperlipidemic mice. J Lipid Res. 2014;55:2073–81. https://doi.org/10.1194/jlr.M049874.
Iacono D, Feltis GC. Impact of apolipoprotein E gene polymorphism during normal and pathological conditions of the brain across the lifespan. Aging. 2019;11(2):787–816. https://doi.org/10.18632/aging.101757ApoE in the context of antagonistic pleiotropy.
Gasparin CC, Leite N, Tureck LV, Souza RLR, Milano-Gai GE, Silva LR, et al. Effects of polymorphisms in APOB, APOE, HSD11β1, PLIN4, and ADIPOQ genes on lipid profile and anthropometric variables related to obesity in children and adolescents. Genet Mol Biol. 2018;41(4):735–41. https://doi.org/10.1590/1678-4685-GMB-2017-0195.
Karjalainen J-P, Mononen N, Hutri-Kähönen N, Lehtimäki M, Hilvo M, Kauhanen M, et al. New evidence from plasma ceramides links apoE polymorphism to greater risk of coronary artery disease in Finnish adults. J Lipid Res. 2019;60:1622–9. https://doi.org/10.1194/jlr.m092809.
Xu M, Zhao J, Zhang Y, Ma X, Dai Q, Zhi H, et al. Apolipoprotein E gene variants and risk of coronary heart disease: a meta-analysis. Biomed Res Int. 2016;3912175:1–12. https://doi.org/10.1155/2016/3912175.
Wolters FJ, Yang Q, Biggs ML, Jakobsdottir J, Li S, Evans DS, et al. The impact of APOE genotype on survival: results of 38,537 participants from six population-based cohorts (E2-CHARGE). PLoS One. 2019;(7):e0219668. https://doi.org/10.1371/journal.pone.0219668Numerous findings about apoE from analysis of a very large database.
Zhang Y, Tang H-Q, Peng W-J, Zhang B-B, Liu M. Meta-analysis for the association of apolipoprotein E ε2/ε3/ε4 polymorphism with coronary heart disease. Chin Med J. 2015;128(10):1391–8.
Bielak LF, Peyser PA. Genetics of subclinical coronary atherosclerosis. Curr Genet Med Rep. 2018;6(3):116–23. https://doi.org/10.1007/s40142-018-0145-xThe impact of genetic variants in subclinical atherosclerosis.
Doliner B, Dong C, Blanton SH, Gardener H, Elkind MSV, Sacco RL, et al. Apolipoprotein E gene polymorphism and subclinical carotid atherosclerosis: the Northern Manhattan Study. Stroke Cerebrovasc Dis. 2018;27(3):645–52. https://doi.org/10.1016/j.jstrokecerebrovasdis.2017.09.053.
Luo J-Q, Ren H, Banh HL, Liu M-Z, Ping Xu P, Fang P-F, et al. The associations between apolipoprotein E gene Epsilon2/Epsilon3/Epsilon4 polymorphisms and the risk of coronary artery disease in patients with type 2 diabetes mellitus. Front Physiol. 2017;8:1031. https://doi.org/10.3389/fphys.2017.01031.
Karschnia P, Nishimura S, Louvi A. Cerebrovascular disorders associated with genetic lesions. Cell Mol Life Sci. 2019;76(2):283–300. https://doi.org/10.1007/s00018-018-2934-5.
Math N, Han TS, Lubomirova I, Hill R, Bentley P, Sharma P. Influences of genetic variants on stroke recovery: a meta-analysis of the 31,895 cases. Neurol Sci. 2019;40:2437–45. https://doi.org/10.1007/s10072-019-04024-w.
Lumsden AL, Mulugeta A, Zhou A, Hyppönen E. Apolipoprotein E (APOE) genotype-associated disease risks: a phenome-wide, registry-based, case-control study utilising the UK Biobank. EbioMedicine. 2020;59:102954. https://doi.org/10.1016/j.ebiom.2020.102954.
Pechlaner R, Tsimikas S, Yin K, Willeit P, Baig P, Santer P, et al. Very-low-density lipoprotein–associated apolipoproteins predict cardiovascular events and are lowered by inhibition of APOC-III. J Am Coll Cardiol. 2017;69(7):790–800. https://doi.org/10.1016/j.jacc.2016.11.065.
Sofat R, Cooper JA, Kumari M, Casas JP, Mitchell JP, Acharya J, et al. Circulating apolipoprotein E concentration and cardiovascular disease risk: metanalysis of results from three studies. PLoS Med. 13(10):e1002146. https://doi.org/10.1371/journal.pmed.1002146.
Rasmussen KL, Tybjærg-Hansen A, Nordestgaard BG, Frikke-Schmidt R. Plasma levels of apolipoprotein E, APOE genotype, and all-cause and cause-specific mortality in 105 949 individuals from a white general population cohort. Eur Heart J. 2019;40:2813–24. https://doi.org/10.1093/eurheartj/ehz402An informative analysis of apoE in a large study.
Valanti E-K, Dalakoura-Karagkouni K, Sanoudou D. Current and emerging reconstituted HDL-apoA-I and HDL-apoE approaches to treat atherosclerosis. J Pers Med. 2018;8:34. https://doi.org/10.3390/jpm8040034.
Koopal C, Marais AD, Westerink J, van der Graaf YT, Visseren FLJ. Effect of adding bezafibrate to standard lipid-lowering therapy on post-fat load lipid levels in patients with familial dysbetalipoproteinemia. A randomized placebo-controlled crossover trial. J Lipid Res. 2017;58:2180–7. https://doi.org/10.1194/jlr.M076901.
Anantharamaiah GM, Garber DW, Goldberg D, Morrel E, Datta G, Palgunachari MN, et al. Novel fatty acyl apoE mimetic peptides have increased potency to reduce plasma cholesterol in mice and macaques. J Lipid Res. 2018;59:2075–83. https://doi.org/10.1194/jlr.M085985.
Gallego-Villar L, Hannibal L, Häberle J, Thöny B, Ben-Omran T, Nasrallah GK, et al. Cysteamine revisited: repair of arginine to cysteine mutations. J Inherit Metab Dis. 2017;40(4):555–67. https://doi.org/10.1007/s10545-017-0060-4.
Ruhaak LR, van der Laarse A, Cobbaert CM. Apolipoprotein profiling as a personalized approach to the diagnosis and treatment of dyslipidaemia. Ann Clin Biochem. 2019;56(3):338–56. https://doi.org/10.1177/0004563219827620.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares no competing interests.
Human and Animal Rights and Informed Consent
The article is a review of publications.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Genetics and Genomics
Rights and permissions
About this article
Cite this article
Marais, A.D. Apolipoprotein E and Atherosclerosis. Curr Atheroscler Rep 23, 34 (2021). https://doi.org/10.1007/s11883-021-00933-4
Accepted:
Published:
DOI: https://doi.org/10.1007/s11883-021-00933-4