Abstract
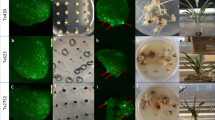
The clustered regulatory interspaced short palindromic repeat/CRISPR-associated protein 9 (CRISPR/Cas9) system has been applied to edit the genomes of quite a few plant species, including perennial woody poplar trees. However, chimeras often exist in primary transgenic plants. For perennial woody trees such as poplar trees, it is difficult to obtain homozygous mutants by self-pollination for due to their long vegetative life and low-seed setting rates. In this study, we report an effective approach to reduce the frequency of chimeric mutants of poplar trees by CRISPR/Cas9 with a second round of shoot regeneration using leaves as the explants. PdbPDS1 was used as the target gene, and only one homozygous PdbPDS1 mutant was obtained from 15 primary transgenic plantlets, which was verified by both the phenotype and the DNA sequence of the PCR product. This indicates that the majority of primary transgenic plantlets in the T0 generation were chimeras. After the second round of shoot regeneration of the chimeric mutants generated by CRISPR/Cas9, approximately 27.0% or 19.1% of the regenerated shoots were homozygous mutants with or without kanamycin selection, respectively. The results showed that a second regeneration could produce homozygous mutant shoots at a high frequency and that kanamycin selection could increase the frequency of homozygous mutant shoots.



Similar content being viewed by others
References
Brooks C, Nekrasov V, Lippman ZB, Van EJ (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol 166:1292–1297
Cermak T, Baltes NJ, Cegan R, Zhang Y, Voytas DF (2015) High frequency, precise modification of the tomato genome. Genome Biol 16:232–245
Chen GQ (2011) Effective reduction of chimeric tissue in transgenics for the stable genetic transformation of Lesquerella fendleri. HortScience 46:86–90
Christou P (1990) Morphological description of transgenic soybean chimeras created by the delivery, integration and expression of foreign DNA using electric discharge particle acceleration. Ann Bot 66:379–386
Domínguez A, Cervera M, Pérez RM, Romero J, Fagoaga C, Cubero J, López MM, Juárez JA, Navarro L, Peńa L (2004) Characterisation of regenerants obtained under selective conditions after Agrobacterium-mediated transformation of citrus explants reveals production of silenced and chimeric plants at unexpected high frequencies. Mol Breed 14:171–183
Dong J, Mchughen A (1993) Transgenic flax plants from Agrobacterium mediated transformation: incidence of chimeric regenerants and inheritance of transgenic plants. Plant Sci 91:139–148
Du H, Zeng H, Zhao M, Cui X, Wang Q, Yang H, Cheng H, Yu D (2016) Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnol 217:90–97
Elorriaga E, Klocko AL, Ma C, Strauss SH (2018) Variation in mutation spectra among CRISPR/Cas9 mutagenized poplars. Front Plant Sci 9:594–605
Faize M, Faize L, Burgos L (2010) Using quantitative real-time PCR to detect chimeras in transgenic tobacco and apricot and to monitor their dissociation. BMC Biotechnol 10:53–60
Fan D, Liu T, Li C, Jiao B, Li S, Hou Y, Luo K (2015) Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci Rep 5:12217
Feng Z et al (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111:4632–4637
Firoozabady E, Deboer DL, Merlo DJ, Halk EL, Amerson LN, Rashka KE, Murray EE (1987) Transformation of cotton (Gossypium hirsutum L.) by Agrobacterium tumefaciens and regeneration of transgenic plants. Plant Mol Biol 10:105–116
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41:e188
Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7:1494–1496
Liang Z, Zhang K, Chen K, Gao C (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics 41:63–68
Maliga P, Nixon PJ (1998) Judging the homoplastomic state of plastid transformants. Trends Plant Sci 3:376–377
Mathews H, Wagoner W, Kellogg J, Bestwick R (1995) Genetic transformation of strawberry: stable integration of a gene to control biosynthesis of ethylene. Vitro Cell Dev Biol Plant 31:36–43
Mathews H, Dewey V, Wagoner W, Bestwick RK (1998) Molecular and cellular evidence of chimaeric tissues in primary transgenics and elimination of chimaerism through improved selection protocols. Transgenic Res 7:123–129
Muruganantham M, Amutha S, Selvaraj N, Vengadesan G, Ganapathi A (2007) Efficient Agrobacterium-mediated transformation of Vigna mungo using immature cotyledonary-node explants and phosphinothricin as the selection agent. Vitro Cell Dev Biol Plant 43:550–557
Osakabe Y, Sugano SS, Osakabe K (2016) Genome engineering of woody plants: past, present and future. J Wood Sci 62:217–225
Schmülling T, Schell J (1993) Transgenic tobacco plants regenerated from leaf disks can be periclinal chimeras. Plant Mol Biol 21:705–708
Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9:2395–2410
Shen Z, Sun J, Yao J, Wang S, Ding M, Zhang H, Qian Z, Zhao N, Sa G, Zhao R, Shen X, Polle A, Chen S (2015) High rates of virus-induced gene silencing by tobacco rattle virus in Populus. Tree Physiol 35:1016–1029
Wang H, Wang C, Liu H, Tang R, Zhang H (2011) An efficient Agrobacterium-mediated transformation and regeneration system for leaf explants of two elite aspen hybrid clones Populus alba × P. berolinensis and Populus davidiana × P. bolleana. Plant Cell Rep 30:2037–2044
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32:947–951
Xing H, Dong L, Wang Z, Zhang H, Han C, Liu B, Wang X, Chen Q (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327–338
Zhou X, Jacobs TB, Xue LJ, Harding SA, Tsai CJ (2015) Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate: CoA ligase specificity and redundancy. New Phytol 208:298–301
Funding
This research was supported the National Natural Science Foundation of China (Grant no. 31800567), the National Key Program on Transgenic Research (Grant no. 2018ZX08020002), the Innovation and Capacity Building Fund in BAAFS (Grant nos. KJCX20170203 and KJCX20180204), the Beijing Natural Science Foundation (Grant no. 6192007), and the National Key R&D Program of China (Grant no. 2016YFD0600104).
Author information
Authors and Affiliations
Contributions
LPD and JHW designed the research. JHW and HZW supervised the experiments. LPD and YJC conducted the experiments. LPD and YM analyzed the results. LPD and JHW drafted the manuscript. JHW and HZW modified the manuscript. All authors reviewed the manuscript.
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ding, L., Chen, Y., Ma, Y. et al. Effective reduction in chimeric mutants of poplar trees produced by CRISPR/Cas9 through a second round of shoot regeneration. Plant Biotechnol Rep 14, 549–558 (2020). https://doi.org/10.1007/s11816-020-00629-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-020-00629-2