Abstract
Gastrointestinal tract disorders, including colorectal cancer (CRC), impose a significant health burden in Europe, with rising incidence rates among both young and elderly populations. Timely detection and removal of polyps, the precursors to CRC, are vital for prevention. Conventional colonoscopy, though effective, is prone to human errors. To address this, we propose an artificial intelligence-based polyp detection system using the YOLO-V8 network. We constructed a diverse dataset from multiple publicly available sources and conducted extensive evaluations. YOLO-V8 m demonstrated impressive performance, achieving 95.6% precision, 91.7% recall, and 92.4% F1-score. It outperformed other state-of-the-art models in terms of mean average precision. YOLO-V8 s offered a balance between accuracy and computational efficiency. Our research provides valuable insights into enhancing polyp detection and contributes to the advancement of computer-aided diagnosis for colorectal cancer.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Gastrointestinal (GI) tract disorders pose a significant public health concern in Europe, leading to approximately 1 million deaths annually. These disorders not only result in substantial mortality, but also impose a considerable burden of illness and healthcare expenses. It is noteworthy that the incidence and prevalence of many GI tract disorders are most prevalent among both the young and, particularly, the elderly population. Given the global aging trend, the burden of these diseases is expected to rise steadily in the future [1].
Colorectal cancer (CRC) is a type of cancer affecting the large intestine and is among the most serious and prevalent forms of cancer. The 5-year survival rates are influenced by various factors and can vary significantly depending on the cancer stage and its location in either the colon or rectum. On average, 5-year survival rates for CRC are estimated to range from 48.6 to 59.4%.[2] Statistics project that by the year 2020, nearly 150,000 individuals will have been diagnosed with CRC, and more than 50,000 will succumb to the disease [3]. Notably, colorectal cancer has experienced the swiftest rise in incidence rates in recent times, with the number of new cases and deaths doubling over the past decade and continuing to increase at an average annual rate of 4–5%. Epidemiological studies reveal a concerning trend of CRC incidence among adults under the age of 50, with the numbers already significantly high and continuing to rise [4]. Research has shown that most CRC cases evolve gradually from colorectal polyps, particularly adenomatous polyps. Timely removal of these polyps through resection can effectively prevent the occurrence of CRC and reduce CRC-related mortality by up to 70% [5]. Addressing this preventive approach is crucial in combatting the increasing prevalence of colorectal cancer.
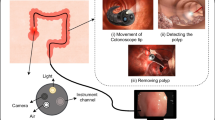
Cancers usually originate as small, non-cancerous growths called polyps, which can eventually develop into cancer (Fig. 1). The adenoma detection rate (ADR) is a key factor in preventing colorectal cancers. When adenomas are detected in time, it can prevent the development of intermediate-stage cancers. Early identification and removal of these growths, commonly known as adenomas, are crucial for effective management. However, there is a wide range in the adenoma detection rate (7–53%) due to individual differences in endoscopists’ technical proficiency.
Benign to malignant progression of colorectal polyps [6]
Polyps are visualized using an invasive technique called colonoscopy, where a camera is inserted into the digestive tract to capture images and identify potential polyps. Colonoscopy has been established as the gold standard for reducing the incidence and mortality of colorectal cancers, as demonstrated in several studies [7, 8]. This imaging procedure has been widely acknowledged as a pivotal method in lowering the incidence of CRC. A study by A.G. Zauber et al. [9] has demonstrated that this imaging approach can lead to a remarkable 53% reduction in mortality by detecting polyps early. Despite the promising results in polyp detection, the procedure itself is susceptible to human errors. The rate of missed polyps during back-to-back colonoscopies can range from approximately 15–30%, depending on the size of the polyp [10].
Studies have identified a significant correlation between the polyp detection rate (PDR) and the ADR, making the PDR a viable alternative index for assessing colonoscopy quality in patients with gastrointestinal diseases [11]. Consequently, it is crucial to address the issue of reducing missed adenomas/polyps through effective means to standardize the quality of colonoscopy, making it a pressing concern in CRC prevention efforts.
In July 2021, Professor Bernal from Barcelona Autonomous University in Spain, a pioneer in the field of computer-aided detection and diagnosis of colorectal polyps, authored the book titled “Computer-Aided Analysis of Gastrointestinal Videos.” This groundbreaking book is the world’s first comprehensive work that compares and analyzes various gastrointestinal image analysis systems. Its primary objective is to support clinicians in completing essential tasks, such as lesion detection in colonoscopy images [12]. Barua et al. conducted a systematic search for the application of artificial intelligence in polyp detection during colonoscopy, using databases like MEDLINE, EMBASE, and Cochrane Central. They compared, summarized, and analyzed the differences between colonoscopy with and without AI by calculating relative risk, absolute risk, and average difference for polyps, adenomas, and colorectal cancer. Their findings revealed that an AI-based polyp detection system can significantly improve the detection rate of non-advanced adenomas and smaller polyps during colonoscopy [13].
1.1 Main contributions
This paper makes a substantial contribution to the field of polyp detection through the innovative application of artificial intelligence, specifically employing the YOLO-V8 methodology. By addressing the critical need for enhanced recognition accuracy and efficiency in polyp detection, this research presents a valuable tool for clinicians to minimize missed diagnoses, facilitate early detection, and contribute to the prevention of colorectal cancers. The introduction of the YOLO-V8 method, with its various iterations (YOLO-V8 n, s, m, l, and x), has been rigorously evaluated across five distinct datasets: Kvasir-SEG [14], CVC-ClinicDB [15], CVC-ColonDB [16], ETIS [17], and EndoScene [18]. Our results, boasting impressive precision, recall, and F1-scores, underscore the efficacy of the approach when compared to other state-of-the-art deep learning-based object detector models.
Beyond its immediate impact, this research also lays a foundational groundwork for future investigations into polyp detection and classification. In a rapidly evolving computer vision landscape, the establishment of a benchmark dataset is of paramount importance. With the aspiration that our dataset will serve as a cornerstone, we anticipate that this work will significantly expedite the progress of computer-aided diagnosis for colorectal cancer. As we delve deeper into the intricate realm of medical image analysis, this paper stands as a pivotal reference point, providing insights and methodologies that can shape and propel future studies in the pursuit of more effective and efficient polyp detection systems.
1.2 Work outline
The organization of the rest of this paper is as follows. In Sect. 2, the related works is explained. Material and applied method are given in Sect. 3. Section 4 explains the experimental results, and Sect. 5 concludes the paper.
2 Related works
2.1 Polyp detection algorithm based on deep learning methods
Various studies have focused on enhancing the detection of polyps during colonoscopy using convolutional neural networks (CNNs). For instance, in [19], a segmentation model based on a three-dimensional, fully convolutional (3D-FCN) network is introduced, achieving state-of-the-art (SOTA) performance in terms of F1-Score and F2-Score. In [20], an FCN network was developed with a unique structure, making initial predictions using binary classification and then processing them through a CNN similar to the U-Net architecture designed in [21]. This network achieved SOTA performance in terms of sensitivity and specificity metrics for the Kvasir-SEG and CVC-ClinicDB datasets. Beyond the challenge of capturing global dependencies, convolutional neural networks (CNNs) face other issues such as overfitting and accurately capturing boundary pixel information. In recent times, efforts have been made to explicitly address these problems, as seen in the development of PraNet [22]. PraNet offers real-time segmentation capabilities through the utilization of deep supervision mechanisms and a reverse attention module for boundary detection. Additionally, the model incorporates a parallel-partial decoder to enhance its performance. These modules have also been implemented in different innovative architectures like AMNet [23]. AMNet further enhances edge-detection capabilities originally employed in PraNet. Recent advancements in polyp segmentation include FANet [24], which presents innovative approaches to attention and refining predictions based on coarser representations. The authors introduced a unique form of attention by leveraging information across training epochs to improve predictions across learnable parameters in subsequent epochs. This attention mechanism proved effective, leading to excellent results when evaluated on the CVC-ClinicDB dataset. However, it did not achieve state-of-the-art performance on the Kvasir-SEG datasets.
Over the past few years, ensemble methods have gained popularity for polyp segmentation, and dual-encoder and/or dual-decoder architectures have emerged [25, 26]. In [25], the dual-encoder–decoder approach demonstrated favorable results in polyp segmentation. However, it applied the dual-model structure sequentially rather than synchronously, with the output of one encoder–decoder serving as input for the following one. Moreover, the network did not introduce many novel components, relying on existing pretrained architectures for its implementation. In [26], a dual-decoder network called DDANet was proposed. It utilized a single ResNet-style encoder with a dual-decoder architecture, generating both a grayscale image and a segmentation mask with each decoder. While this approach showcased creativity, subsequent works have produced significant improvements in the metrics generated by the network. Indeed, in pursuit of improving the accuracy of output segmentation maps for polyp detection, various ensemble methods, particularly dual-model approaches, have been explored. Examples of such approaches include the dual mask R-CNN model [27] and the combination of dual DeiT transformer and ResNet CNN structure proposed in [28].
To enhance the efficiency of polyp detection, several target detection algorithms based on the YOLO series have been developed. Guo et al. presented an automatic polyp detection algorithm utilizing the YOLO-V3 structure combined with active learning. This approach effectively reduces the false positive rate in polyp detection [29]. Cao et al. introduced a feature extraction and fusion module, integrating it with the YOLO-V3 network. By incorporating both high-level and low-level feature maps, this method can capture semantic information and outperforms other techniques in detecting small polyps [30]. Pacal et al. proposed a real-time automatic polyp detection method based on YOLO-V4. They integrated the cspnet network into the architecture and incorporated the mish activation function, Diou loss function, and transformer block. This approach demonstrates higher accuracy and superior performance compared to previous methods [31]. These advancements in target detection algorithms based on the YOLO series hold promise for more efficient and accurate polyp detection during colonoscopy. Lee and colleagues [32] introduced a real-time system for polyp detection utilizing YOLO-V4. The system employed a multiscale mesh for the identification of small polyps. Performance enhancements were achieved through the incorporation of advanced data augmentation techniques and the utilization of different activation functions. Wan and colleagues [12] proposed a model based on YOLO-V5 for real-time polyp detection, incorporating a self-attention mechanism. With this approach, the method strengthens beneficial features, weakens less relevant ones, resulting in an enhanced performance of polyp detection. In a comprehensive experimental study, Pacal et al. [33] assessed novel datasets, SUN and PICCOLO, using the Scaled YOLO-V4 algorithm. The results of the experimental studies indicate that the SUN and PICCOLO datasets demonstrate exceptional success in polyp detection, with the Scaled YOLO-V4 algorithm standing out as one of the most suitable object detection algorithms for large-scale datasets. Durak and colleagues [34] conducted training on state-of-the-art object detection algorithms, including YOLO-V4 [35], CenterNet, EfficientDet [36], and YOLO-V3 [37], for automatic gastric polyp detection. In the experimental results, the YOLO-V4 algorithm demonstrated the highest performance compared to other methods, showcasing its effectiveness for deployment in CAD systems for automatic polyp detection. Qian et al. [38] proposed a method that combines GAN architectures with the YOLO-V4 object detection algorithm for robust polyp detection. Experimental evaluations were carried out on three publicly available datasets. The results indicated that the proposed method outperforms U-Net, synthesizes more realistic polyp images and significantly improves polyp detection performance. Gabriel provides a comprehensive implementation of YOLO-V4 at various precision levels (FP32, FP16, and INT8) for polyp detection. Notably, the study explores the previously untested INT8 quantization level. The research employs Darknet for YOLO-V4 training, integrates TensorRT for quantization and optimization (FP16 and INT8), and evaluates different data augmentation and regularization techniques on benchmark datasets of Etis-Larib and CVC-ClinicDB. The study achieves commendable performance metrics, including a median average precision (mAP) of 82.93% for Etis-Larib and 90.96% for CVC-ClinicDB. The analysis encompasses GPU specifications, inference speeds, and accuracy metrics for each precision level, revealing potential regularization effects with quantization. The findings contribute valuable insights to the application of quantization, particularly in the context of larger and more complex models like YOLO-V4, strengthening the argument for its role in network regularization [39]. In [40] by Ahmet Karaman and Ishak Pacal, the study introduces a groundbreaking integration of the ABC algorithm with YOLO-based object detection algorithms, focusing on optimizing activation functions and hyperparameters—an unprecedented exploration in the literature. This integration achieves more efficient optimization in a single operation, saving time and hardware costs. Key contributions include a 3% improvement in real-time polyp detection performance with the YOLO-V5 algorithm, marking the first demonstration of real-time capabilities on SUN and PICCOLO datasets. The study thoroughly examines the influence of activation functions and hyperparameters on real-time polyp detection accuracy. The proposed method is versatile, easily applicable to any dataset and YOLO-based algorithm, tailoring parameters to optimize performance [41].
As the resolution of polyp images in colonoscopy increases, the feature representation of these images becomes more complex, comprising a large number of pixels. Traditional polyp detection methods often fail to effectively preprocess these intricate features from the original images. During the polyp detection process, various challenges arise due to inherent characteristics of colorectal images, such as low brightness, presence of noise, reduced contrast, and technical limitations of imaging equipment. These factors can result in blurred edges between adjacent tissues, leading to difficulties in extracting optimal features for subsequent analysis. Moreover, the demand for real-time polyp detection has grown significantly. Despite comprehensive research conducted on automatic polyp detection systems over the past decade, there remains a lack of evidence regarding the system’s ability to accurately locate and track polyps during real-time colonoscopy in clinical practice. Additionally, researchers must continue to explore and develop real-time polyp detection systems to ensure their practical viability and efficacy in clinical settings.
3 Materials and methods
3.1 Datasets
The model’s training and evaluation process involved the use of a total of five publicly available datasets. Specifically, the datasets used for evaluation were Kvasir-SEG [14], CVC-ClinicDB [15], CVC-ColonDB [16], ETIS [17], and EndoScene [18].
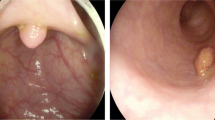
To validate the performance of the applied method, the model underwent training and testing procedures. The training dataset was a combination of all five datasets, totaling 1890 images. The model’s performance was then tested on the remaining 10% of unused data from the Kvasir-SEG and CVC-ClinicDB datasets, as well as the benchmark polyp datasets: ETIS, CVC-ColonDB, and EndoScene. A validation set was created using a 10% subsample of the training data. The allocation of images from the Kvasir-SEG, CVC-ClinicDB, CVC-ColonDB, ETIS, and EndoScene datasets into training, testing, and validation sets followed a random selection process based on the proportions mentioned above. More details regarding these divisions can be found in Table 1, and some samples of our datasets are presented in Fig. 2.
In this study, we adopted an approach to augment our dataset, customizing the augmentation method to suit the sensitive nature of medical images. The augmentation process involved various operations, including normalization, brightness adjustment, and hue augmentation. To maintain the integrity of the medical data, we applied conservative alterations to the images, ensuring that only minor changes were introduced.
3.2 Overall architecture of the method
Our primary objective in the detection strategy centers on enhancing the detection capabilities, with a particular emphasis on polyp in colonoscopy images. To achieve this, we utilize YOLO-V8 [42], an improved iteration of the original YOLO [43]. YOLO-V8 has attained state-of-the-art performance through optimizations in model structure, anchor box or anchor-free schemes, and the implementation of diverse data augmentation techniques. Our deep learning architecture is based on the five different sized versions of YOLO-V8. The YOLO-V8 framework offers five distinct models N, S, M, L, X, each characterized by varying channel depth and filter numbers. For the backbone architecture, we opted to use all five models due to its balanced combination of detection accuracy and processing speed.
One of the primary advantages of incorporating YOLO-V8 into the computer vision project is its enhanced accuracy compared to previous YOLO models. YOLO-V8 offers support for multiple tasks, such as object detection, instance segmentation, and image classification, enhancing its versatility for various applications. YOLO-V8 represents the most recent advancement in the YOLO object detection model, with a primary focus on enhancing both accuracy and efficiency compared to its predecessors. Key updates in this iteration comprise an optimized network architecture, a redesigned anchor box implementation, and a modified loss function, all contributing to a notable boost in overall detection precision. YOLO-V8 has showcased enhanced accuracy when compared to its earlier iterations, positioning it as a strong competitor alongside state-of-the-art object detection models. Designed with efficiency in mind, YOLO-V8 is optimized to run smoothly on standard hardware, making it a practical and viable choice for real-time object detection tasks, including edge computing scenarios. Anchor boxes are used in YOLO-V8 to match predicted bounding boxes to ground truth bounding boxes, improving the overall accuracy of the object detection process.
3.2.1 Backbone
YOLO-V8 ’s training process is expected to be notably faster in comparison with two-stage object detection models, making it an efficient choice for projects requiring rapid training times. In comparison with ultralytics/YOLO-V5 [44], the backbone of the system experienced modifications with the replacement of C3 by C2f and integrating the ELAN concept from YOLO-V7 [45]. Specifically, the first 6x6 convolution in the stem was replaced with a 3x3 convolution. This integration enhances the model’s capacity to acquire more comprehensive gradient flow information. The C3 module is composed of three ConvModules and n DarknetBottleNecks, while the C2f module incorporates two ConvModules and n DarknetBottleNecks connected through Split and Concat. The ConvModule is structured with Conv-BN-SiLU, and ’n’ denotes the quantity of bottlenecks. Additionally, in C2f, the outputs from the Bottleneck, which comprises two 3x3 convolutions with residual connections, are combined, while in C3, only the output from the last Bottleneck was used. Two convolutions (#10 and #14 in the YOLO-V5 config) were removed from the YOLO-V8 configuration. The bottleneck in YOLO-V8 remains the same as in YOLO-V5, except for the change in the first convolution’s kernel size from 1x1 to 3x3. This modification indicates a shift toward the ResNet block as defined in 2015.
3.2.2 Head
In contrast to the YOLO-V5 model, which employs a coupled head, the approach incorporates a decoupled head, separating the classification and detection heads. The model eliminates the objectness branch, retaining only the classification and regression branches. Anchor-Base utilizes numerous anchors in the image to ascertain the four offsets of the regression object from the anchors, refining the object’s precise location with the aid of corresponding anchors and offsets. The architecture of the model is shown in Fig. 3.
YOLO-V8: model architecture including backbone and head [42]
3.2.3 Loss
In the model training, we employ the Task Aligned Assigner from Task-aligned One-stage Object Detection (TOOD) [46] for the assignment of positive and negative samples. This assigner selects positive samples by considering the weighted scores of classification and regression, as represented in Eq. 1.
Here, \(s\) represents the predicted score associated with the labeled class, and \(u\) denotes the Intersection over Union (IoU) between the prediction and the ground truth bounding box. Moreover, the model incorporates classification and regression branches. The classification branch utilizes binary cross-entropy (BCE) loss, as depicted by the following equation:
In this context, \(w\) represents the weight, \(y_n\) is the labeled value, and \(x_n\) is the predicted value generated by the model.
For the regression branch, we employ distribute focal loss (DFL) [47] and complete IoU (CIoU) loss [48]. DFL is applied to broaden the probability distribution around the object \(y\), and its equation is expressed as follows:
Here, the equations for \(S_n\) and \(S_{n+1}\) are presented below:
\(\hbox {CIoU}_\textrm{Loss}\) incorporates an influential factor into distance IoU (DIoU) loss [49], taking into account the aspect ratio of both the prediction and the ground truth bounding box. The equation is as follows:
Here, \(n\) represents the parameter quantifying the aspect ratio’s consistency, and its definition is provided as follows:
4 Results and discussion
4.1 Evaluation metrics
This paper assesses the algorithm’s performance for polyp detection using four indicators: precision, recall, and F1-score. The formulas for these indicators are as follows:
Among these indicators, TP represents the count of true positives, indicating the number of correctly detected and labeled polyp instances. FN represents the count of false negatives, referring to the number of polyps that were not correctly detected. FP stands for the count of false positives, representing the number of non-polyp regions misclassified as polyps.
Precision assesses the ratio of correctly labeled polyps among all predicted polyp instances and serves as a metric that measures the percentage of correct predictions. In the context of polyp detection, it indicates the confidence level when a positive detection is made. A higher precision value helps in reducing the occurrence of false alarms, which can alleviate financial and mental stress for clients. Recall, on the other hand, represents the fraction of detected objects. In polyp detection, this metric holds significant importance since a higher recall ensures that more patients receive timely further checks and appropriate treatment. Consequently, it can lead to reduced mortality and prevent excessive costs for patients. It measures the proportion of polyps detected among all polyp images. The F1-score is a combined metric that takes both precision and recall into account. By considering both false positives and false negatives, it provides a balanced assessment of a model’s performance. It serves as the harmonic mean of precision and recall, offering a comprehensive evaluation of the algorithm’s performance.
4.2 Performance of the applied method
The hyperparameter configurations are detailed in Table 2. The evaluation of each model’s performance relied on three metrics: precision, recall, and F1-score. All the results shown in Table 2 highlight the impressive performance of the YOLO-V8 variations in polyp detection tasks in our test dataset.
As a concluding step, we evaluated the effectiveness of our applied method on the datasets used in this project. We compared the results obtained using our applied method with those from other existing methods with the same datasets. The comparative outcomes are presented in Table 3. This table showcases the performance of our applied method in relation to the alternative approaches, providing valuable insights into its effectiveness and potential advantages.
Visual results of the applied method, along with the available detected polyp with bounding boxes, are presented in Fig. 4.
In the realm of colorectal polyp detection, the YOLO-V8 m model stands out as a formidable contender, surpassing various state-of-the-art models in terms of recall, precision, and F1-score, as delineated in Table 3. Noteworthy achievements include outperforming custom architectures like Tajbakhsh et al.’s (2015b) by a significant margin, showcasing a 91.2% recall, 95.1% precision, and a 91.4% F1-score. In comparison with YOLO-V1 (Zheng et al., 2018), YOLO-V8 m consistently demonstrates superior results, notably achieving 91.2% recall, 95.1% precision, and a 91.4% F1-score on CVC-ClinicDB, 90.7% recall, 94.6% precision, and a 94.4% F1-score on ETIS, and 91.4% recall, 94.4% precision, and a 92.1% F1-score on CVC-ColonDB. Even against hybrid architectures like Urban et al.’s (2018) ResNet-50, VGG16, and VGG19, our model maintains competitive performance across diverse datasets. Moreover, when compared to innovative designs such as Zhang et al.’s [53] single-shot multibox detector and other YOLO versions, YOLO-V8 m consistently demonstrates a robust balance between accuracy and computational efficiency. This comprehensive evaluation positions YOLO-V8 m as an advanced and reliable solution for real-time colorectal polyp detection, contributing substantial insights to the ongoing evolution of artificial intelligence in medical imaging applications.
YOLO-V8 incorporates an efficient backbone architecture that enables streamlined information flow. The model’s design optimally captures intricate features relevant to polyp detection while minimizing unnecessary computational burden. This efficiency ensures real-time processing without compromising accuracy. YOLO-V8 emphasizes attention to various scales and resolutions within the input image. The model effectively addresses the multiscale nature of polyps, allowing it to discern details at different levels. This adaptability contributes significantly to achieving higher precision, recall, and F1-scores in comparison with larger, less flexible models. YOLO-V8 leverages advanced training strategies and data augmentation techniques. The model is adept at learning from diverse datasets, which is particularly crucial in polyp detection where variations in size, shape, and appearance are common. This adaptability enhances generalization and robustness, resulting in improved performance on unseen data. YOLO-V8 benefits from the evolutionary improvements introduced in YOLO-V5 and builds upon them. The modifications and changes implemented in YOLO-V5 to achieve YOLO-V8 play a pivotal role in refining the model’s accuracy. By acknowledging and incorporating these advancements, YOLO-V8 surpasses the limitations of earlier versions and outperforms larger, more resource-intensive models.
4.3 Limitations and future directions
The presented methodology signifies a significant stride beyond previous iterations of YOLO algorithms, showcasing improved efficacy in optimizing hyperparameters with considerations for both time and cost. However, the study’s broader implications were hindered by the limited availability of an extensive public polyp dataset. Despite achieving favorable results, certain datasets from the literature were excluded due to their scant polyp images and a restricted number of patients. This underscores the inherent data dependency of deep learning algorithms, highlighting the critical need for robust datasets to showcase optimal performance. Noteworthy advancements are observed in real-time speed and detection efficiency, surpassing existing methodologies and prior YOLO versions. Current endeavors aim to extend these methods to clinical applications by amalgamating existing datasets. Nevertheless, the recognition of the necessity for datasets featuring a more diverse array of polyp images, representing various patients and geographic locations, is acknowledged. Contemplation of additional studies and forthcoming research endeavors is underway, foreseeing the development of more effective models for clinical applications through the utilization of larger and more diverse datasets in the future.
5 Conclusion
Currently, artificial intelligence polyp detection technology is in its early stages of development. When compared to traditional statistics or expert systems, deep learning methods typically exhibit notable enhancements in performance and detection accuracy for most image target detection tasks. In response to this problem, the focus of this article revolves around polyp target detection, specifically employing YOLO-V8 as the chosen method. In this article, we have successfully created a relatively extensive endoscopic dataset specifically designed for detecting polyps in colonoscopy images. In our comprehensive exploration of artificial intelligence polyp detection technology, the utilization of YOLO-V8, particularly the YOLO-V8 m variant, has emerged as a standout performer. The success of YOLO-V8 m, with a precision of 95.6%, recall of 91.7%, and an F1-score of 92.4%, can be attributed to a judicious balance between its range of parameters and the characteristics of our extensive endoscopic dataset tailored for polyp detection in colonoscopy images. The meticulous design of our dataset, emphasizing images with low contrasts, enabled YOLO-V8 m to significantly enhance detection accuracy, particularly in challenging scenarios. The model’s proficiency is further underscored by its notable mean average precision at 50% overlap (mAP50) of 85.4% and mAP50-95 of 62%. Moreover, the inference time of 10.6 milliseconds and 25 million parameters demonstrate a commendable equilibrium between accuracy and computational efficiency, rendering YOLO-V8 m a compelling choice for real-time applications. While YOLO-V8 m stands out as a prime choice, YOLO-V8 s, with a precision of 95.2%, recall of 90.5%, and F1-score of 91.7%, also showcases commendable performance. The nuanced trade-off between accuracy and computational efficiency is evident in its slightly longer inference time of 4.7 milliseconds and 11 million parameters. This research can establish a fundamental reference point for future studies concerning polyp detection and classification. Considering the rapid progress in the computer vision domain over the past years, the presence of a benchmark dataset holds paramount importance. It is our aspiration that our dataset will substantially expedite the computer-aided diagnosis of colorectal cancer.
Data availability
All relevant datasets used in this study will be made available upon request.
References
Sahafi, A., Wang, Y., Rasmussen, C., Bollen, P., Baatrup, G., Blanes-Vidal, V., Herp, J., Nadimi, E.: Edge artificial intelligence wireless video capsule endoscopy. Sci. Rep. 12(1), 13723 (2022)
Lewis, J., Cha, Y.-J., Kim, J.: Dual encoder–decoder-based deep polyp segmentation network for colonoscopy images. Sci. Rep. 13(1), 1183 (2023)
Siegel, R.L., Miller, K.D., Goding Sauer, A., Fedewa, S.A., Butterly, L.F., Anderson, J.C., Cercek, A., Smith, R.A., Jemal, A.: Colorectal cancer statistics, 2020. CA Cancer J. Clin. 70(3), 145–164 (2020)
Stoffel, E.M., Murphy, C.C.: Epidemiology and mechanisms of the increasing incidence of colon and rectal cancers in young adults. Gastroenterology 158(2), 341–353 (2020)
Kudo, S.-E., Mori, Y., Misawa, M., Takeda, K., Kudo, T., Itoh, H., Oda, M., Mori, K.: Artificial intelligence and colonoscopy: current status and future perspectives. Digest. Endosc. 31(4), 363–371 (2019)
healthline: https://www.healthline.com/health/colorectal-cancer/colon-polyp-size-chart#screening-guidelines (2023)
Bibbins-Domingo, K., Grossman, D.C., Curry, S.J., Davidson, K.W., Epling, J.W., García, F.A., Gillman, M.W., Harper, D.M., Kemper, A.R., Krist, A.H., et al.: Screening for colorectal cancer: us preventive services task force recommendation statement. JAMA 315(23), 2564–2575 (2016)
Rex, D.K., Boland, C.R., Dominitz, J.A., Giardiello, F.M., Johnson, D.A., Kaltenbach, T., Levin, T.R., Lieberman, D., Robertson, D.J.: Colorectal cancer screening: recommendations for physicians and patients from the us multi-society task force on colorectal cancer. Gastroenterology 153(1), 307–323 (2017)
Zauber, A.G., Winawer, S.J., O’Brien, M.J., Lansdorp-Vogelaar, I., Ballegooijen, M., Hankey, B.F., Shi, W., Bond, J.H., Schapiro, M., Panish, J.F., et al.: Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N. Engl. J. Med. 366(8), 687–696 (2012)
Matsuda, T., Ono, A., Kakugawa, Y., Matsumoto, M., Saito, Y.: Impact of screening colonoscopy on outcomes in colorectal cancer. Jpn. J. Clin. Oncol. 45(10), 900–905 (2015)
Ng, S., Sreenivasan, A.K., Pecoriello, J., Liang, P.S.: Polyp detection rate correlates strongly with adenoma detection rate in trainee endoscopists. Digest. Dis. Sci. 65, 2229–2233 (2020)
Wan, J., Chen, B., Yu, Y.: Polyp detection from colorectum images by using attentive yolov5. Diagnostics 11(12), 2264 (2021)
Barua, I., Vinsard, D.G., Jodal, H.C., Løberg, M., Kalager, M., Holme, Ø., Misawa, M., Bretthauer, M., Mori, Y.: Artificial intelligence for polyp detection during colonoscopy: a systematic review and meta-analysis. Endoscopy 53(03), 277–284 (2020)
Ro, Y.M., Cheng, W.-H., Kim, J., Chu, W.-T., Cui, P., Choi, J.-W., Hu, M.-C., De Neve, W.: MultiMedia Modeling: 26th International Conference, MMM 2020, Daejeon, South Korea, January 5–8, 2020, Proceedings, Part II, vol. 11962. Springer, Berlin (2019)
Bernal, J., Sánchez, F.J., Fernández-Esparrach, G., Gil, D., Rodríguez, C., Vilariño, F.: Wm-dova maps for accurate polyp highlighting in colonoscopy: validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 43, 99–111 (2015)
Tajbakhsh, N., Gurudu, S.R., Liang, J.: Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans. Med. Imaging 35(2), 630–644 (2015)
Silva, J., Histace, A., Romain, O., Dray, X., Granado, B.: Toward embedded detection of polyps in wce images for early diagnosis of colorectal cancer. Int. J. Comput. Assist. Radiol. Surg. 9, 283–293 (2014)
Vázquez, D., Bernal, J., Sánchez, F.J., Fernández-Esparrach, G., López, A.M., Romero, A., Drozdzal, M., Courville, A., et al.: A benchmark for endoluminal scene segmentation of colonoscopy images. J. Healthc. Eng. 2017, 66 (2017)
Yu, L., Chen, H., Dou, Q., Qin, J., Heng, P.A.: Integrating online and offline three-dimensional deep learning for automated polyp detection in colonoscopy videos. IEEE J. Biomed. Health Inform. 21(1), 65–75 (2016)
Pozdeev, A.A., Obukhova, N.A., Motyko, A.A.: Automatic analysis of endoscopic images for polyps detection and segmentation. In: 2019 IEEE Conference of Russian Young Researchers in Electrical and Electronic Engineering (EIConRus), pp. 1216–1220. IEEE (2019)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III, vol. 18, pp. 234–241. Springer, Berlin (2015)
Fan, D.-P., Ji, G.-P., Zhou, T., Chen, G., Fu, H., Shen, J., Shao, L.: Pranet: parallel reverse attention network for polyp segmentation. In: International Conference on Medical Image Computing and Computer-assisted Intervention, pp. 263–273. Springer, Berlin (2020)
Song, P., Li, J., Fan, H.: Attention based multi-scale parallel network for polyp segmentation. Comput. Biol. Med. 146, 105476 (2022)
Tomar, N.K., Jha, D., Riegler, M.A., Johansen, H.D., Johansen, D., Rittscher, J., Halvorsen, P., Ali, S.: Fanet: a feedback attention network for improved biomedical image segmentation. IEEE Trans. Neural Netw. Learn. Syst. 6, 66 (2022)
Galdran, A., Carneiro, G., Ballester, M.A.G.: Double encoder–decoder networks for gastrointestinal polyp segmentation. In: Pattern Recognition. ICPR International Workshops and Challenges: Virtual Event, January 10–15, 2021, Proceedings, Part I, pp. 293–307. Springer, Berlin (2021)
Tomar, N.K., Jha, D., Ali, S., Johansen, H.D., Johansen, D., Riegler, M.A., Halvorsen, P.: Ddanet: dual decoder attention network for automatic polyp segmentation. In: Pattern Recognition. ICPR International Workshops and Challenges: Virtual Event, January 10–15, 2021, Proceedings, Part VIII, pp. 307–314. Springer, Berlin (2021)
Kang, J., Gwak, J.: Ensemble of instance segmentation models for polyp segmentation in colonoscopy images. IEEE Access 7, 26440–26447 (2019)
Zhang, Y., Liu, H., Hu, Q.: Transfuse: fusing transformers and cnns for medical image segmentation. In: Medical Image Computing and Computer Assisted Intervention—MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part I 24, pp. 14–24. Springer, Berlin (2021)
Guo, Z., Zhang, R., Li, Q., Liu, X., Nemoto, D., Togashi, K., Niroshana, S.I., Shi, Y., Zhu, X.: Reduce false-positive rate by active learning for automatic polyp detection in colonoscopy videos. In: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI). IEEE, pp. 1655–1658 (2020)
Cao, C., Wang, R., Yu, Y., Zhang, H., Yu, Y., Sun, C.: Gastric polyp detection in gastroscopic images using deep neural network. PLoS ONE 16(4), 0250632 (2021)
Pacal, I., Karaboga, D.: A robust real-time deep learning based automatic polyp detection system. Comput. Biol. Med 134, 104519 (2021)
Lee, J.-n, Chae, J.-w, Cho, H.-c: Improvement of colon polyp detection performance by modifying the multi-scale network structure and data augmentation. J. Electr. Eng. Technol. 17(5), 3057–3065 (2022)
Pacal, I., Karaman, A., Karaboga, D., Akay, B., Basturk, A., Nalbantoglu, U., Coskun, S.: An efficient real-time colonic polyp detection with yolo algorithms trained by using negative samples and large datasets. Comput. Biol. Med. 141, 105031 (2022)
Durak, S., Bayram, B., Bakırman, T., Erkut, M., Doğan, M., Gürtürk, M., Akpınar, B.: Deep neural network approaches for detecting gastric polyps in endoscopic images. Med. Biol. Eng. Comput. 59, 1563–1574 (2021)
Liu, W., Anguelov, D., Erhan, D., Szegedy, C., Reed, S., Fu, C.-Y., Berg, A.C.: Ssd: Single shot multibox detector. In: Computer Vision—ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, October 11–14, 2016, Proceedings, Part I 14, pp. 21–37. Springer, Berlin (2016)
Tan, M., Pang, R., Le, Q.V.: Efficientdet: scalable and efficient object detection. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 10778–10787 (2020). https://doi.org/10.1109/CVPR42600.2020.01079
Bochkovskiy, A., Wang, C.-Y., Liao, H.-Y.M.: Yolov4: optimal speed and accuracy of object detection. arXiv preprint arXiv:2004.10934 (2020)
Qian, Z., Jing, W., Lv, Y., Zhang, W.: Automatic polyp detection by combining conditional generative adversarial network and modified you-only-look-once. IEEE Sens. J. 22(11), 10841–10849 (2022)
Carrinho, P., Falcao, G.: Highly accurate and fast yolov4-based polyp detection. Available at SSRN 4227573 (2022)
Karaman, A., Karaboga, D., Pacal, I., Akay, B., Basturk, A., Nalbantoglu, U., Coskun, S., Sahin, O.: Hyper-parameter optimization of deep learning architectures using artificial bee colony (abc) algorithm for high performance real-time automatic colorectal cancer (crc) polyp detection. Appl. Intell. 53(12), 15603–15620 (2023)
Karaman, A., Pacal, I., Basturk, A., Akay, B., Nalbantoglu, U., Coskun, S., Sahin, O., Karaboga, D.: Robust real-time polyp detection system design based on yolo algorithms by optimizing activation functions and hyper-parameters with artificial bee colony (abc). Expert Syst. Appl. 221, 119741 (2023)
Ultralytics: https://github.com/ultralytics/ultralytics (2013)
Redmon, J., Divvala, S., Girshick, R., Farhadi, A.: You only look once: unified, real-time object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 779–788 (2016)
Ge, Z., Liu, S., Wang, F., Li, Z., Sun, J.: Yolox: Exceeding yolo series in 2021. arXiv preprint arXiv:2107.08430 (2021)
Wang, C.-Y., Bochkovskiy, A., Liao, H.-Y.M.: Yolov7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 7464–7475 (2023)
Feng, C., Zhong, Y., Gao, Y., Scott, M.R., Huang, W.: Tood: task-aligned one-stage object detection. In: 2021 IEEE/CVF International Conference on Computer Vision (ICCV), pp. 3490–3499. IEEE Computer Society (2021)
Li, X., Wang, W., Wu, L., Chen, S., Hu, X., Li, J., Tang, J., Yang, J.: Generalized focal loss: learning qualified and distributed bounding boxes for dense object detection. Adv. Neural Inf. Process. Syst. 33, 21002–21012 (2020)
Zheng, Z., Wang, P., Ren, D., Liu, W., Ye, R., Hu, Q., Zuo, W.: Enhancing geometric factors in model learning and inference for object detection and instance segmentation. IEEE Trans. Cybernet. 52(8), 8574–8586 (2021)
Zheng, Z., Wang, P., Liu, W., Li, J., Ye, R., Ren, D.: Distance-iou loss: faster and better learning for bounding box regression. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 34, pp. 12993–13000 (2020)
Tajbakhsh, N., Gurudu, S.R., Liang, J.: Automatic polyp detection in colonoscopy videos using an ensemble of convolutional neural networks. In: 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), pp. 79–83 (2015). IEEE
Zheng, Y., Zhang, R., Yu, R., Jiang, Y., Mak, T.W., Wong, S.H., Lau, J.Y., Poon, C.C.: Localisation of colorectal polyps by convolutional neural network features learnt from white light and narrow band endoscopic images of multiple databases. In: 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 4142–4145. IEEE 2018)
Urban, G., Tripathi, P., Alkayali, T., Mittal, M., Jalali, F., Karnes, W., Baldi, P.: Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology 155(4), 1069–1078 (2018)
Zhang, X., Chen, F., Yu, T., An, J., Huang, Z., Liu, J., Hu, W., Wang, L., Duan, H., Si, J.: Real-time gastric polyp detection using convolutional neural networks. PLoS ONE 14(3), 0214133 (2019)
Wang, D., Zhang, N., Sun, X., Zhang, P., Zhang, C., Cao, Y., Liu, B.: Afp-net: realtime anchor-free polyp detection in colonoscopy. In: 2019 IEEE 31st International Conference on Tools with Artificial Intelligence (ICTAI), pp. 636–643 (2019). IEEE
Lee, J.Y., Jeong, J., Song, E.M., Ha, C., Lee, H.J., Koo, J.E., Yang, D.-H., Kim, N., Byeon, J.-S.: Real-time detection of colon polyps during colonoscopy using deep learning: systematic validation with four independent datasets. Sci. Rep. 10(1), 8379 (2020)
Qadir, H.A., Shin, Y., Solhusvik, J., Bergsland, J., Aabakken, L., Balasingham, I.: Toward real-time polyp detection using fully cnns for 2d gaussian shapes prediction. Med. Image Anal. 68, 101897 (2021)
Xu, J., Zhao, R., Yu, Y., Zhang, Q., Bian, X., Wang, J., Ge, Z., Qian, D.: Real-time automatic polyp detection in colonoscopy using feature enhancement module and spatiotemporal similarity correlation unit. Biomed. Signal Process. Control 66, 102503 (2021)
Liu, X., Guo, X., Liu, Y., Yuan, Y.: Consolidated domain adaptive detection and localization framework for cross-device colonoscopic images. Med. Image Anal. 71, 102052 (2021)
Nogueira-Rodríguez, A., Domínguez-Carbajales, R., Campos-Tato, F., Herrero, J., Puga, M., Remedios, D., Rivas, L., Sánchez, E., Iglesias, A., Cubiella, J., et al.: Real-time polyp detection model using convolutional neural networks. Neural Comput. Appl. 34(13), 10375–10396 (2022)
Li, Q., Yang, G., Chen, Z., Huang, B., Chen, L., Xu, D., Zhou, X., Zhong, S., Zhang, H., Wang, T.: Colorectal polyp segmentation using a fully convolutional neural network. In: 2017 10th International Congress on Image and Signal Processing, Biomedical Engineering and Informatics (CISP-BMEI), pp. 1–5. IEEE (2017)
Tashk, A., Herp, J., Nadimi, E., Sdu, S.U.: Automatic segmentation of colorectal polyps based on a novel and innovative convolutional neural network approach. WSEAS Tran, Syst. Control 14, 384–391 (2019)
Qadir, H.A.: Development of image processing algorithms for the automaticscreening of colon cancer. PhD thesis, University of Oslo, Norway (2020)
Akbari, M., Mohrekesh, M., Nasr-Esfahani, E., Soroushmehr, S.R., Karimi, N., Samavi, S., Najarian, K.: Polyp segmentation in colonoscopy images using fully convolutional network. In: 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 69–72. IEEE (2018)
Funding
Open access funding provided by University of Southern Denmark We would like to clarify that this research received no specific funding from any organization or institution. The study was self-funded by the authors.
Author information
Authors and Affiliations
Contributions
Both authors have contributed equally.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lalinia, M., Sahafi, A. Colorectal polyp detection in colonoscopy images using YOLO-V8 network. SIViP 18, 2047–2058 (2024). https://doi.org/10.1007/s11760-023-02835-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-023-02835-1