Abstract
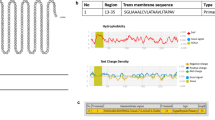
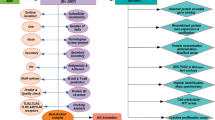
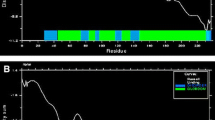
Mycobacterium tuberculosis, the etiological agent of tuberculosis, is one of the trickiest pathogens. We have only a few protective shields, like the BCG vaccine against the pathogen, which itself has poor efficacy in preventing adult tuberculosis. Even though different vaccine trials for an alternative vaccine have been conducted, those studies have not shown much promising results. In the current study, advanced computational technology was used to study the potential of a novel hypothetical mycobacterial protein, identified by subtractive hybridization, to be a vaccine candidate. NHP2 (Novel Hypothetical Protein 2), housed in the RD7 region of the clinical strains of M. tuberculosis, was studied for its physical, chemical, immunological and structural properties using different computational tools. PFAM studies and Gene ontology studies depicted NHP2 protein to be functionally active with a possible antibiotic binding domain too. Different computational tools used to assess the toxicity, allergenicity and antigenicity of the protein indicated its antigenic nature. Immune Epitope Database (IEDB) tools were used to study the T and B cell determinants of the protein. The 3D structure of the protein was designed, refined and authenticated using bioinformatics tools. The validated tertiary structure of the protein was docked against the TLR3 immune receptor to study the binding affinity and docking scores. Molecular dynamic simulation of the protein-protein complex formed were studied. NHP2 was found to activate host immune response against tubercle bacillus and could be explored as a potential vaccine in the fight against tuberculosis.










Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. Nucleotide sequence is available in NCBI; GenBank Accession Number—GU994138.2.
Abbreviations
- BCG:
-
Bacillus Calmette Guerin
- NHP2:
-
Novel Hypothetical Protein 2
- IEDB:
-
Immune Epitope Database
- TLR3:
-
Toll Like receptor
- TB:
-
Tuberculosis
- MDR TB:
-
Multi Drug Resistant Tuberculosis
- RD:
-
Region of Differences
- ESAT:
-
Early Secreted Antigenic Target
- RvDs:
-
Rv Deletions
- NHP1:
-
Novel Hypothetical Protein 1
- NCBI:
-
National Center for Biotechnology Information
- ORF:
-
Open Reading Frames
- EMBL:
-
European Molecular Biology Laboratory
- SMS:
-
Sequence Manipulation Suite
- CDD:
-
Conserved Domain Search
- BLAST:
-
Basic Local Alignment Search Tool
- SMART:
-
Simple Modular Architecture Research Tool
- PFAM:
-
Protein Family Database
- CELLO:
-
SubCELlular LOcalization prediction
- TMHMM:
-
Trans Membrane Hidden Markov Model
- PFP FunDSeq:
-
Predicting Protein Fold Pattern with Functional Domain and Sequential Evolution Information
- CTL:
-
Cytotoxic T Lymphocyte
- HLA:
-
Human Leukocytes Antigen
- MHC:
-
Major Histocompatibility Complex
- pMHC:
-
Peptide MHC
- PSIPRED:
-
PSI-blast based secondary structure PREDiction
- SOPMA:
-
Self-Optimized Prediction Method with Alignment
- NMR:
-
Nuclear magnetic Resonance
- PDB:
-
Protein Data Bank
- MD:
-
Molecular Dynamic Simulation
- NMA:
-
Normal Mode Analysis
- Tet-R:
-
Tetracyclin Repressor
- NK cells:
-
Natural Killer cells
- RMSD:
-
Root Mean Square Deviation
- 3-D:
-
3 Dimensional
References
Al Tbeishat H (2022) Novel In Silico mRNA vaccine design exploiting proteins of M. tuberculosis that modulates host immune responses by inducing epigenetic modifications. Sci Rep 12(1):4645. https://doi.org/10.1038/s41598-022-08506-4
Albutti A (2021) An integrated computational framework to design a multi-epitopes vaccine against Mycobacterium tuberculosis. Sci Rep 11:21929. https://doi.org/10.1038/s41598-021-01283-6
Al-Megrin WAI, Karkashan A, Alnuqaydan AM, Aba Alkhayl FF, Alrumaihi F, Almatroudi A, Allemailem KS (2022) Design of a multi-epitopes based chimeric vaccine against Enterobacter cloacae using pan-genome and reverse vaccinology approaches. Vaccines 10(6):886. https://doi.org/10.3390/vaccines10060886
Andreatta M, Karosiene E, Rasmussen M, Stryhn A, Buus S, Nielsen M (2015) Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics 67(11):641–650. https://doi.org/10.1007/s00251-015-0873-y
Andrusier N, Nussinov R, Wolfson HJ (2007) FireDock: fast interaction refinement in molecular docking. Proteins 69(1):139–59. https://doi.org/10.1002/prot.21495
Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, Durbin R (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res 29(1):37–40. https://doi.org/10.1093/nar/29.1.37
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA (2000) Gene ontology: tool for the unification of biology. Nat Genet 25(1):25–29. https://doi.org/10.1038/75556
Bai W, Liu H, Ji Q, Zhou Y, Liang L, Zheng R, Chen J, Liu Z, Yang H, Zhang P, Kaufmann SH, Ge B (2014) TLR3 regulates mycobacterial RNA-induced IL-10 production through the PI3K/AKT signaling pathway. Cell Signal 26(5):942–950. https://doi.org/10.1016/j.cellsig.2014.01.015
Bhasin M, Raghava GP (2004) SVM based method for predicting HLA-DRB1* 0401 binding peptides in an antigen sequence. Bioinform 20(3):421–3. https://doi.org/10.1093/bioinformatics/btg424
Bibi S, Ullah I, Zhu B, Adnan M, Liaqat R, Kong WB, Niu S (2021) In silico analysis of epitope-based vaccine candidate against tuberculosis using reverse vaccinology. Sci Rep 11(1):1–6. https://doi.org/10.1038/s41598-020-80899-6
Blanco JRL, Aliaga JI, Quintana-Ortí ES, Chacón P (2014) iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res 1(42):271–276. https://doi.org/10.1093/nar/gku339
Buchan DW, Jones DT (2019) The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res 47:W402–W407. https://doi.org/10.1093/nar/gkz297
Calis JJ, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, Sette A, Keşmir C, Peters B (2013) Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol 9(10):1003266. https://doi.org/10.1371/journal.pcbi.1003266
Coppola M, Ottenhoff TH (2018) Genome wide approaches discover novel Mycobacterium tuberculosis antigens as correlates of infection, disease, immunity and targets for vaccination. Seminars in immunology 1 (Vol. 39, pp. 88–101). Academic Press. https://doi.org/10.1016/j.smim.2018.07.001
Danilchanka O, Mailaender C, Niederweis M (2008) Identification of a novel multidrug efflux pump of Mycobacterium tuberculosis. Antimicrobial Agents Chemother 52(7):2503–2511. https://doi.org/10.1128/AAC.00298-08
Dimitrov I, Flower DR, Doytchinova I (2013) AllerTOP-a server for in silico prediction of allergens. BMC Bioinform 14(6):1–9. https://doi.org/10.1186/1471-2105-14-S6-S4
Dimitrov I, Naneva L, Doytchinova I, Bangov I (2014) AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinform 30(6):846–851. https://doi.org/10.1093/bioinformatics/btt619
Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform 8(1):1–7. https://doi.org/10.1186/1471-2105-8-4
Duhovny D, Nussinov R, Wolfson HJ (2002) Efficient unbound docking of rigid molecules. In: International workshop on algorithms in bioinformatics 17 (pp. 185–200). Springer, Berlin. https://doi.org/10.1007/3-540-45784-4_14
Eisenberg D, Lüthy R, Bowie JU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Meth Enzymo 277:396–404. https://doi.org/10.1038/356083a0
Ejalonibu MA, Ogundare SA, Elrashedy AA, Ejalonibu MA, Lawal MM, Mhlongo NN, Kumalo HM (2021) drug discovery for mycobacterium tuberculosis using structure-based computer-aided drug design approach. Int J Mol Sci 22(24):13259. https://doi.org/10.3390/ijms222413259
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M (2014) Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. https://doi.org/10.1093/nar/gkt1223
Garg A, Gupta D (2008) VirulentPred: a SVM based prediction method for virulent proteins in bacterial pathogens. BMC Bioinform 9(1):1–2. https://doi.org/10.1186/1471-2105-9-62
Garnier J, Gibrat JF, Robson B (1996) GOR method for predicting protein secondary structure from amino acid sequence. Meth Enzymo 266:540–553. https://doi.org/10.1016/s0076-6879(96)66034-0
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (eds) The proteomics protocols handbook. Springer Protocols Handbooks. Humana Press. https://doi.org/10.1385/1-59259-890-0:571
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinform 11(6):681–684. https://doi.org/10.1093/bioinformatics/11.6.681
Global tuberculosis report (2020) Geneva: World Health Organization; 2020. Licence: CC BY-NC-SA 3.0 IGO
Gomez M, Johnson S, Gennaro ML (2000) Identification of secreted proteins of Mycobacterium tuberculosis by a bioinformatic approach. Infect Immun 68(4):2323–2327. https://doi.org/10.1128/IAI.68.4.2323-2327.2000
Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Raghava GP (2013) In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 8(9):73957. https://doi.org/10.1371/journal.pone.0073957
Hirokawa T, Boon-Chieng S, Mitaku S (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinform 14(4):378–379. https://doi.org/10.1093/bioinformatics/14.4.378
Ikai A (1980) Thermostability and aliphatic index of globular proteins. J Biochem 88(6):1895–1898
Jendele L, Krivak R, Skoda P, Novotny M, Hoksza D (2019) PrankWeb: a web server for ligand binding site prediction and visualization. Nucleic Acids Res 2(47):345–349. https://doi.org/10.1093/nar/gkz424
Jespersen MC, Peters B, Nielsen M, Marcatili P (2017) BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res 45:W24–W29. https://doi.org/10.1093/nar/gkx346
Jespersen MC, Mahajan S, Peters B, Nielsen M, Marcatili P (2019) Antibody specific B-cell epitope predictions: leveraging information from antibody-antigen protein complexes. Front Iimmunol 26(10):298. https://doi.org/10.3389/fimmu.2019.00298
Ji Z, Jian M, Chen T, Luo L, Li L, Dai X, Bai R, Ding BY, Wen S, Zhou G, Abi ME, Liu A, Bao F (2019) Immunogenicity and safety of the M72/AS01E candidate vaccine against tuberculosis: a meta-analysis. Front Immunol 3(10):2089. https://doi.org/10.3389/fimmu.2019.02089
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596(7873):583–589. https://doi.org/10.1038/s41586-021-03819-2
Kaufmann SHE (2020) Vaccination against tuberculosis: revamping BCG by molecular genetics guided by immunology. Front Immunol 27(11):316. https://doi.org/10.3389/fimmu.2020.00316
Khan Z, Ualiyeva D, Amissah OB, Sapkota S, Hameed HM, Zhang T (2022) Insight into novel Anti-tuberculosis vaccines by using immunoinformatics approaches. Front Microbiol 13:866873. https://doi.org/10.3389/fmicb.2022.866873
Kim DE, Chivian D, Baker D (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32(2):W526–W531. https://doi.org/10.1093/nar/gkh468
Ko J, Park H, Heo L, Seok C (2012) GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res 40(W1):W294–W297. https://doi.org/10.1093/nar/gks493
Kootery KP, Sarojini S (2022) Structural and functional characterization of a hypothetical protein in the RD7 region in clinical isolates of Mycobacterium tuberculosis—an in silico approach to candidate vaccines. J Genet Eng Biotechnol 20(1):1–5. https://doi.org/10.1186/s43141-022-00340-5
Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S (2017) The ClusPro web server for protein–protein docking. Nat Protoc 12(2):255–278. https://doi.org/10.1038/nprot.2016.169
Krivák R, Hoksza D (2018) P2Rank: machine learning based tool for rapid and accurate prediction of ligand binding sites from protein structure. J Cheminformatics 10(1):1–2. https://doi.org/10.1186/s13321-018-0285-8
Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M (2007) Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform 8(1):1–2. https://doi.org/10.1186/1471-2105-8-424
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26(2):283–291. https://doi.org/10.1107/S0021889892009944
Letunic I, Khedkar S, Bork P (2021) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49(D1):D458–D460. https://doi.org/10.1093/nar/gkaa937
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48(D1):D265–D268. https://doi.org/10.1093/nar/gkz991
Machado D, Lecorche E, Mougari F, Cambau E, Viveiros M. (2018) Insights on mycobacterium leprae efflux pumps and their implications in drug resistance and virulence. Front Microbiol 3072. https://doi.org/10.3389/fmicb.2018.03072
Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32(Web Server issue):W327–W331. https://doi.org/10.1093/nar/gkh454
Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ (2008) FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res 19(36):W229-32. https://doi.org/10.1093/nar/gkn186
Mirzayev F, Viney K, Linh NN, Gonzalez-Angulo L, Gegia M, Jaramillo E, Zignol M, Kasaeva T (2021) World Health Organization recommendations on the treatment of drug-resistant tuberculosis. Eur Respir J 57(6):2003300. https://doi.org/10.1183/13993003.03300-2020
Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer E, Tosatto S, Paladin L, Raj S, Richardson LJ, Finn RD (2021) Bateman A. Pfam: The protein families database in 2021. Nucleic Acids Res. 49(D1):D412–D419. https://doi.org/10.1093/nar/gkaa913
Möller S, Croning MDR, Apweiler R (2001) Evaluation of methods for the prediction of membrane spanning regions. Bioinform 17(7):646–653. https://doi.org/10.1093/bioinformatics/17.7.646
Mustafa AS (2021) Immunological Characterization of Proteins Expressed by Genes Located in Mycobacterium tuberculosis-Specific Genomic Regions Encoding the ESAT6-like Proteins. Vaccines 9(1):27. https://doi.org/10.3390/vaccines9010027
Nemes E, Geldenhuys H, Rozot V, Rutkowski KT, Ratangee F, Bilek N, Mabwe S, Makhethe L, Erasmus M, Toefy A, Mulenga H, Hanekom WA, Self SG, Bekker LG, Ryall R, Gurunathan S, DiazGranados CA, Andersen P, Kromann I, Evans T, Ellis RD, Landry B, Hokey DA, Hopkins R, Ginsberg AM, Scriba TJ, Hatherill M C-040-404 Study Team (2018) Prevention of M tuberculosis infection with H4:IC31 vaccine or BCG revaccination. N Engl J Med 379(2):138–149. https://doi.org/10.1056/NEJMoa1714021
Nguyen H, Gazy N, Venketaraman V (2020) A role of intracellular toll-like receptors (3, 7, and 9) in response to mycobacterium tuberculosis and co-infection with HIV. Int J Mol Sci 21(17):6148. https://doi.org/10.3390/ijms21176148
Oso BJ, Oyewo EB, Oladiji AT (2021) Homology modelling and analysis of structure predictions of human tumour necrosis factor ligand superfamily member 8. Future J Pharm Sci 7(1):1–2. https://doi.org/10.1186/s43094-021-00262-y
Park T, Baek M, Lee H, Seok C (2019) GalaxyTongDock: Symmetric and asymmetric ab initio protein–protein docking web server with improved energy parameters. J Comput Chem 40(27):2413–7. https://doi.org/10.1002/jcc.25874
Passi A, Rajput NK, Wild DJ, Bhardwaj A (2018) RepTB: a gene ontology based drug repurposing approach for tuberculosis. J Cheminformatics 10(1):1–2. https://doi.org/10.1186/s13321-018-0276-9
Petersen TN, Brunak S, Von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8(10):785–6. https://doi.org/10.1038/nmeth.1701
Ponting CP, Schultz J, Milpetz F, Bork P (1999) SMART: identification and annotation of domains from signalling and extracellular protein sequences. Nucleic Acids Res 27(1):229–232. https://doi.org/10.1093/nar/27.1.229
Prabhakaran K, Harris EB, Randhawa B (1999) Bactericidal action of ampicillin/sulbactam against intracellular mycobacteria. Int J Aantimicrob Agents 13(2):133–5. https://doi.org/10.1016/s0924-8579(99)00101-6
Priya VG, Shanmuga MU, Mehta M (2013) Function and structure prediction of Rv2004c, a hypothetical protein from M. tuberculosis. Int J Innov Res Sci Eng Technol 2:38–46
Rahman O, Cummings SP, Harrington DJ, Sutcliffe IC (2008) Methods for the bioinformatic identification of bacterial lipoproteins encoded in the genomes of Gram-positive bacteria. World J Microbiol Biotechnol 24(11):2377–2382. https://doi.org/10.1099/00221287-148-7-2065
Rao D, Vasanthapuram R, Satishchandra P, Desai A (2018) Pattern recognition receptor mRNA expression and cytokine and granzyme levels in HIV infected individuals with neurotuberculosis. J Neuroimmunol 318:21–28. https://doi.org/10.1016/j.jneuroim.2018.01.015
Rashid M, Saha S, Raghava GP (2007) Support Vector Machine-based method for predicting subcellular localization of mycobacterial proteins using evolutionary information and motifs. BMC Bioinform 13(8):337. https://doi.org/10.1186/1471-2105-8-337
Saha S, Raghava GPS (2004) BcePred: Prediction of continuous b-cell epitopes in antigenic sequences using physico-chemical properties. In: Nicosia G, Cutello V, Bentley PJ, Timmis J (eds) Artificial Immune Systems. ICARIS 2004. Lecture Notes inComputer Science, vol 3239. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-540-30220-9_16
Saha S, Raghava GPS (2006) Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins 65(1):40–48. https://doi.org/10.1002/prot.21078
Saikat AS, Kabir ML, Khalipha AB (2020) An In silico approach for structural and functional annotation of uncharacterized protein Rv0986 present in mycobacterium tuberculosis. Eur J Med Health Sci 2(3):61–67. https://doi.org/10.3390/proceedings2020066013
Šali A, Potterton L, Yuan F, van Vlijmen H, Karplus M (1995) Evaluation of comparative protein modeling by MODELLER. Proteins 23(3):318–326. https://doi.org/10.1002/prot.340230306
Sarojini S, Mundayoor S (2020) An ancestral genomic locus in Mycobacterium tuberculosis clinical isolates from India hints the genetic link with Mycobacterium canettii. Int Microbiol 23(3):397–404. https://doi.org/10.1007/s10123-019-00113-0
Sarojini S, Soman S, Radhakrishnan I, Mundayoor S (2005) Identification of moa A3 gene in patient isolates of Mycobacterium tuberculosis in Kerala, which is absent in M. tuberculosis H37Rv and H37Ra. BMC Infect Dis 5(1):1–7. https://doi.org/10.1186/1471-2334-5-81
Sarojini S, Madhavilatha GK, Soman S, Ajay Kumar R, Mundayoor S (2011) A novel site of insertion of IS6110 in the moaB3 gene of a clinical isolate of Mycobacterium tuberculosis. Microbiol Res 2(1):26–29. https://doi.org/10.4081/mr.2011.e7
Shen HB, Chou KC (2009) Predicting protein fold pattern with functional domain and sequential evolution information. J Theor Biol 256(3):441–446. https://doi.org/10.1016/j.jtbi.2008.10.007
Soman S, Joseph BV, Sarojini S, Kumar RA, Katoch VM, Mundayoor S (2007) Presence of region of difference 1 among clinical isolates of Mycobacterium tuberculosis from India. J Clin Microbiol 45(10):3480–3481. https://doi.org/10.1128/JCM.01234-07
Stothard P (2000) The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28(6):1102–1104. https://doi.org/10.2144/00286ir01
Tariq MH, Bhatti R, Ali NF, Ashfaq UA, Shahid F, Almatroudi A, Khurshid M (2021) Rational design of chimeric multiepitope Based Vaccine (MEBV) against human T-cell lymphotropic virus type 1: An integrated vaccine informatics and molecular dockingbased approach. PLoS ONE 16(10):0258443. https://doi.org/10.1371/journal.pone.0258443
Vangone A, Bonvin AM (2015) Contacts-based prediction of binding affinity in protein–protein complexes. elife 20(4):e07454. https://doi.org/10.7554/eLife.07454
Weng G, Wang E, Wang Z, Liu H, Zhu F, Li D, Hou T (2019) HawkDock: a web server to predict and analyze the protein–protein complex based on computational docking and MM/GBSA. Nucleic Acids Res 47(W1):W322-30. https://doi.org/10.1093/nar/gkz397
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35(2):W407–W410. https://doi.org/10.1093/nar/gkm290
Yu CS, Lin CJ, Hwang JK (2004) Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci 13(5):1402–1406. https://doi.org/10.1110/ps.03479604
Zaks K, Jordan M, Guth A, Sellins K, Kedl R, Izzo A, Bosio C, Dow S (2006) Efficient immunization and cross-priming by vaccine adjuvants containing TLR3 or TLR9 agonists complexed to cationic liposomes. J Immunol 176:7335–7345. https://doi.org/10.4049/jimmunol.176.12.7335
Funding
The research work was supported by the Karnataka Science and Technology Academy (KSTA), Govt of Karnataka through the ShortTerm Research Grant.
Author information
Authors and Affiliations
Contributions
SS conceived the idea and KPK performed the experimental parts. Both authors were involved in data analysis, interpretation and writing the article. Both authors have read the final draft of the article.
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kootery, K.P., Sarojini, S. In silico analysis of NHP2 membrane protein, a novel vaccine candidate present in the RD7 region of Mycobacterium tuberculosis. Biologia 79, 355–372 (2024). https://doi.org/10.1007/s11756-023-01559-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11756-023-01559-4