Abstract
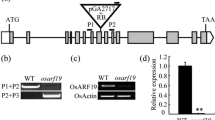
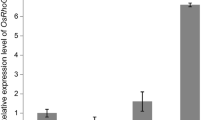
The transition of meristems is an important developmental process for crop plants. Florigen is considered to be produced in leaves, then moves into the shoot apical meristem (SAM), triggers the transition from the vegetative to the reproductive phase. However, little is known whether Florigen functions in callus development or not. By fused reporter gene β-glucuronidase (GUS) to 1.7 kb promoter of Heading date 3a (Hd3a), GUS signals were detected in the scutellum cells, as well as in green point of the putative transgenic calli. Quantitative RT-PCR results demonstrated that the expression level of Hd3a was increased gradually over time along with the transition from scutellum-deprived callus to shoot. As reported that ectopic expression of FT-like genes caused earlier flowering, we also found that 80% constitutive expression of Hd3a transgenic callus showed formation floral-like organ structures. However, Hd3a RNA interference (RNAi) transgenic calli did not show any obvious phenotype, although AP1 or AP1-like genes—OsMADS14, OsMADS15, and OsMADS18- expression level is decreased during callus development. Both in Hd3a and RFT1 overexpression transgenic calli, Hd3a also modulated AP1 or AP1-like genes, as well AEPALLATA (SEP)-like gene, OsMADS34 during green point formation. Meanwhile, transgenic calli of RFT1and OsMADS50, but not OsEhd1, shared similar results as Hd3a. All of these findings suggested that florigen genes Hd3a and RFT1 have partial conserved functions in the transition of meristems during callus development.






Similar content being viewed by others
References
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309:1052–1056
Ahn JH (2016) Flowering time: have florigen, will travel. Nat Plants 2:16081
An G, Ebert PR, Mitra A, Ha SB (1988) Binary vectors. Plant molecular biology manual, vol A3. Kluwer Academic Publishers, Dortrecht, pp 1–19
Cho LH, Yoon J, Pasriga R, An G (2016) Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol 170:2159–2171
Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long distance signaling in floral induction of Arabidopsis. Science 316:1030–1033
Fornara F, Parenicova L, Falasca G, Pelucchi N, Masiero S, Ciannamea S, Lopez-Dee Z, Altamura MM, Colombo L, Kater MM (2004) Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol 135:2207–2219
Gao XC, Liang WQ, Yin CS, Ji SM, Wang HM, Su XA, Guo CC, Kong HZ, Xue HW, Zhang DB (2010) The SEPALLATA-Like geneOsMADS34is required for rice inflorescence and spikelet development. Plant Physiol 153:728–740
He C, Chen X, Huang H, Xu L (2012) Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet 8(8):e1002911
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hu Y, Liang W, Yin CS, Yang XJ, Ping B, Li A, Jia R, Chen MJ, Luo ZJ, Cai Q et al (2015) Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol Plant 8:1366–1384
Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11(11):550–558
Jeon JS, Lee S, Jung K, Yang W, Yi G, Oh B, An G (2000) Production of transgenic rice plants showing reduced heading date and plant height by ectopic expression of rice MADS-box genes. Mol Breed 6:581–592
Kaneko-Suzuki M, Kurihara-Ishikawa R, Okushita-Terakawa C, Kojima C, Nagano-Fujiwara M, Ohki I, Tsuji H, Shimamoto K, Taoka KI (2018) TFL1-like proteins in rice antagonize rice FT-like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant Cell Physiol 59:458–468
Kim SR, Lee DY, Yang JI, Moon S, An G (2009) Cloning vectors for rice. J Plant Biol 52:73–78
Kim SL, Choi M, Jung KH, An G (2013) Analysis of the early-flowering mechanisms and generation of T-DNA tagging lines in Kitaake, a model rice cultivar. J Exp Bot 64:4169–4182
Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, Yoshida H, Nagamura Y, Kyozuka J (2012) Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24(5):1848–1859
Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135(4):767–774
Komiya R, Yokoi S, Shimamoto K (2009) A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136:3443–3450
Lee YS, An G (2015) OsGI controls flowering time by modulating rhythmic flowering time regulators in rice preferentially under short day. J Plant Biol 58:137–145
Lee S, Jeon J, Jung K, An G (1999) Binary vectors for efficient transformation of rice. J Plant Biol 42:310–316
Lee S, Kim J, Han JJ, Han MJ, An G (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J 38:754–764
Pasriga R, Cho LH, Yoon JM, An G (2018) Identification of the regulatory region responsible for vascular tissue-specific expression in the rice Hd3a promoter. Mol Cells 41(4):342–350
Purwestri YA, Oqaki Y, Tamaki S, Tsuji H, Shimamoto K (2009) The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant Cell Physiol 50:429–438
Putterill J, Varkonyi-Gasic E (2016) FT and florigen long-distance flowering control in plants. Curr Opin Plant Biol 33:77–82
Ryu CH, Lee S, Cho LH et al (2009) OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ 32:1412–1427
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316:1033–1036
Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S et al (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476(7360):332–335
Tsuji H (2017) Molecular function of florigen. Breed Sci 67(4):327–332
Tsuji H, Tachibana C, Tamaki S, Taoka K, Kyozuka J, Shimamoto K (2015) Hd3a promotes lateral branching in rice. Plant J 82(2):256–266
Wei JH, Choi HB, Jin P, Wu YF, Yoon J, Lee YS, Quan TY, An G (2016) GL2-type homeobox gene Roc4 in rice promotes flowering time preferentially under long days by repressing Ghd7. Plant Sci 252:133–143
Wei JH, Wu YF, Cho LH, Yoon JM, Choi HB, Jin P, Yi JK, Lee YS, Jeong HJ, Yang JI, An G (2017) Identification of root-preferential transcription factors in rice by analyzing GUS expression patterns of T-DNA tagging lines. J Plant Biol 60(3):268–277
Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309:1056–1059
Xu L (2017) De novo root regeneration from leaf explants: wounding, auxin, and cell fate transition. Curr Opin Plant Biol 41:39–45
Yang J, Lee S, Hang R, Kim SR, Lee YS, Cao X, Amasino R, An G (2013) OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J 73:566–578
Yun D, Liang W, Dreni L, Yin C, Zhou Z, Kater MM, Zhang D (2013) OsMADS16 genetically interacts with OsMADS3 and OsMADS58 in specifying floral patterning in rice. Mol Plant 6:743–756
Zhu YJ, Fan YY, Wang K, Huang DR, Liu WZ, Ying JZ, Zhuang JY (2017) Rice Flowering Locus T 1 plays an important role in heading date influencing yield traits in rice. Sci Rep 7(1):4918
Acknowledgements
This study was supported by the Natural Science Foundation of China (31571573, 31701351), a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and Yangzhou University Postdoctoral Science Foundation. We thank Gynheung An and Kyungsook An in KyungHee university for providing the suggestions in this project and managing the seed stock, respectively.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Additional information
Communicated by J. Huang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, Y., Wei, J., Choi, SC. et al. Rice florigen gene Hd3a has conserved functions in callus development. Acta Physiol Plant 41, 125 (2019). https://doi.org/10.1007/s11738-019-2914-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-019-2914-x