Abstract
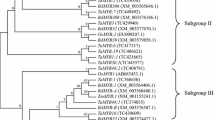
Myeloblastosis (MYB) proteins constitute one of the largest transcription factor (TF) families in plants and play crucial roles in regulating plant physiological and biochemical processes, including adaptation to diverse abiotic stresses. These TF families contain highly conserved MYB repeats (1R, R2R3, 3R and 4R) at the N-terminus. Roles for MYB TFs have been reported in response to such stresses as dehydration, salt, cold, and drought. The characterization of Masson pine (Pinus massoniana) MYB TFs are reported, including the analysis of MYB TFs expression in seedlings under controlled conditions and two different phosphate (Pi) deficient treatments. By searching for conserved MYB motifs in full transcriptomic RNA sequencing data for P. massoniana, 59 sequences were identified as MYB TFs. Conserved domain structures and comparative functional and phylogenetic relationships of these MYB TFs with those in Arabidopsis were assessed using various bioinformatics tools. Based on microarray analysis, P. massoniana MYB genes exhibited different expression patterns under the two Pi deficiency conditions. Genes encoding MYB TFs that showed increased expression under critical Pi deficiency were identified, and some MYBs were differentially expressed only under conditions of severe Pi starvation. These results are useful for the functional characterization of MYB TFs that may be involved in the response to Pi deficiency and play divergent roles in plants.





Similar content being viewed by others
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281:37636–37645
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Baek D, Kim MC, Chun HJ, Kang S, Park HC, Shin G, Park J, Shen M, Hong H, Kim WY, Kim DH, Lee SY, Bressan RA, Bohnert HJ, Yun DJ (2013) Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol 161(1):362–373
Cai H, Tian S, Dong H (2012) Large scale in silico identification of MYB family genes from wheat expressed sequence tags. Mol Biotechnol 52(2):184–192
Dai X, Wang Y, Yang A, Zhang WH (2012) OsMYB2P-1, and R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiol 159:169–183
Denekamp M, Smeekens SC (2003) Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol 132:1415–1423
Devaiah BN, Madhuvanthi R, Athikkattuvalasu R, Karthikeyan S, Raghothama KG (2009) Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant 2:43–58
Ding Z, Li S, An X, Liu X, Qin H, Wang D (2009) Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics 36:17–29
Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, Tang YX (2012) Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol 12:106
Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55(6):940–953
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15(10):573–581
Fan F, Cui B, Zhang T, Qiao G, Ding G, Wen X (2014) The temporal transcriptomic response of Pinus massoniana seedlings to phosphorus deficiency. PLoS ONE 9(8):e105068
Feng S, Xu Y, Yang L, Sun S, Wang D, Chen X (2015) Genome-wide identification and characterization of R2R3-MYB transcription factors in pear. Sci Hortic 197:176–182
Ganesan G, Sankararamasubramanian HM, Harikrishnan M, Ganpudi A, Parida A (2012) A MYB transcription factor from the grey mangrove is induced by stress and confers NaCl tolerance in tobacco. J Exp Bot 63:4549–4561
Ito M (2005) Conservation and diversification of three-repeat Myb transcription factors in plants. J Plant Res 118:61–69
Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41:577–585
Jin JP, Tian F, Yang DC, Meng YQ, Kong L, Luo JC, Gao G (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res D45(11):D1040–D1045
Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom 13(1):544
Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411(6838):706–709
Kim JH, Nguyen NH, Jeong CY, Nguyen NT, Hong SW, Lee H (2013) Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J Plant Physiol 170(6):1461–1465
Klempnauer K, Gonda T, Bishop J (1982) Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell 31:453–463
Kobayashi K, Suzuki T, Iwata E, Nakamichi N, Suzuki T, Chen P, Ohtani M, Ishida T, Hosoya H, Müller S, Leviczky T, Pettkó-Szandtner A, Darula Z, Iwamoto A, Nomoto M, Tada Y, Higashiyama T, Demura T, Doonan JH, Hauser MT, Sugimoto K, Umeda M, Magyar Z, Bögre L, Ito M (2015) Transcription repression by MYB3R proteins regulates plant organ growth. EMBO J 34:1992–2007
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40(1):302–305
Liao W, Yang Y, Li Y, Wang G, Peng M (2016) Genome-wide identification of cassava R2R3 MYB family genes related to abscission zone separation after environmental stress-induced abscission. Sci Rep 6:32006
Lotkowska ME, Tohge T, Fernie AR, Xue GP, Balazadeh S, Mueller-Roeber B (2015) The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol 169:1862–1880
Matus JT, Aquea F, Arce-Johnson P (2008) Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol 8:83
Mehra P, Pandey BK, Giri J (2017) Improvement in phosphate acquisition and utilization by a secretory purple acid phosphatase (OsPAP21b) in rice. Plant Biotechnol J. https://doi.org/10.1111/pbi.12699
Muthamilarasan M, Bonthala VS, Khandelwal R, Jaishankar J, Shweta S, Nawaz K, Prasad M (2015) Global analysis of WRKY transcription factor superfamily in Setaria identifies potential candidates involved in abiotic stress signaling. Front Plant Sci 6:910
Ogata K, Hojo H, Aimoto S, Nakai T, Nakamura H, Sarai A, Ishii S, Nishimura Y (1992) Solution structure of a DNA-binding unit of MYB: a helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc Natl Acad Sci USA 89:6428–6432
Pandey BK, Mehra P, Verma L, Bhadouria J, Giri J (2017) OsHAD1, a haloacid dehalogenase-like APase enhances phosphate accumulation. Plant Physiol. https://doi.org/10.1104/pp.17.00571
Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J 6:3553–3558
Peng X, Liu H, Wang D, Shen S (2016) Genome-wide identification of the Jatrophacurcas MYB family and functional analysis of the abiotic stress responsive gene JcMYB2. BMC Genom 17:251
Ren F, Guo QQ, Chang LL, Chen L, Zhao CZ, Zhong H, Li XB (2012) Brassica napus PHR1 gene encoding a MYB-like protein functions in response to phosphate starvation. PLoS ONE 7(8):e44005
Rose A, Meier I, Wienand U (1999) The tomato I-box binding factor LeMYBI is a member of a novel class of MYB-like proteins. Plant J 20:641–652
Rosinski JA, Atchley WR (1998) Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol 46:74–83
Ruan W, Guo M, Wu P, Yi K (2017) Phosphate starvation induced OsPHR4 mediates Pi-signaling and homeostasis in rice. Plant Mol Biol 93(3):327–340
Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15(6):2122–2133
Seo PJ, Park CM (2009) Auxin homeostasis during lateral root development under drought condition. Plant Signal Behav 4:1002–1004
Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23:1138–1152
Shen J, Yan L, Zhang J, Li H, Bai Z, Chen X, Zhang W, Zhang F (2011) Phosphorus dynamics: from soil to plant. Plant Physiol 156(3):997–1005
Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J (2007) Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev Biol 311(2):566–578
Stracke R, Holtgräwe D, Schneider J, Pucker B, Sörensen TR, Weisshaar B (2014) Genome-wide identification and characterization of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol 14(1):249
Su L, Wang Y, Liu D, Li X, Zhai Y, Sun X, Li X, Liu Y, Li J, Wang Q (2015) The soybean gene, GmMYNJ2, encodes a R2R3-type transcription factor involved in drought stress tolerance in Arabidopsis thaliana. Acta Physiol Plant 37:138
Tombuloglu H, Kekec G, Sakcali MS, Unver T (2013) Transcriptome-wide identification of R2R3-MYB transcription factors in barley with their boron responsive expression analysis. Mol Genet Genomics 288:141–155
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homologs is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529–1539
Wang J, Sun J, Miao J, Guo J, Shi Z, He M, Chen Y, Zhao X, Li B, Han F, Tong Y, Li Z (2013) A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signaling and increases grain yield in wheat. Ann Bot 111:1139–1153
Wang F, Suo Y, Wei H, Li M, Xie C, Wang L, Chen X, Zhang Z (2015) Identification and characterization of 40 isolated Rehmannia glutinosa MYB family genes and their expression profiles in response to shading and continuous cropping. Int J Mol Sci 16:15009–15030
Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21(4):1109–1128
Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM (2009) Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol 149(2):981–993
Yang A, Dai XY, Zhang WH (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bor 63:2541–2556
Zhang Y, Zhou Z, Ma X, Jin G (2010) Foraging ability and growth performance of four subtropical tree species in response to heterogeneous nutrient environments. J For Res 15:91–98
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Project funding: This study was funded by the Natural Science Foundation of Guizhou Province, P. R. China (20161051), the National Natural Science Foundation of China (31660185) and the Doctoral Fund Project of Guizhou University, P. R. China (201464).
The online version is available at http://www.springerlink.com
Corresponding editor: Yu Lei.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11676_2019_911_MOESM5_ESM.tif
Supplementary Fig. S2 Conserved amino acid residues are present among the 59 proteins of the P. massoniana MYBs. The amino acid sequences are aligned, and gaps (dots) have been introduced to maximize the alignment (TIFF 3687 kb)
Rights and permissions
About this article
Cite this article
Fan, F., Wang, Q., Wen, X. et al. Transcriptome-wide identification and expression profiling of Pinus massoniana MYB transcription factors responding to phosphorus deficiency. J. For. Res. 31, 909–919 (2020). https://doi.org/10.1007/s11676-019-00911-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-019-00911-2