Abstract
Background
The MYB family is one of the most abundant transcription factor families in plants. MYB proteins are involved in plant development, abiotic stress tolerance, hormone signal transduction and disease resistance. Here we perform genome-wide identification of MYB family transcription factors in an energy plant J. curcas, including determining family composition, phylogenetic evolution and functional prediction analysis. In addition, we further elucidate the function of the JcMYB2 gene.
Methods
The phylogenetic trees were constructed by using the neighbor-joining method in MEGA 5.2. The biological functions of some JcMYBs were predicted according to orthology. The full length cDNA of JcMYB2 was cloned by using the RACE method. GUS histochemical staining was used to test the activity of the JcMYB2 promoter. Expression patterns of JcMYB2 were detected by using qPCR Transcriptional activity JcMYB2 were confirmed through yeast one hybrid. Subcellular Localization of JcMYB2 Protein were demonstrated by transient expression in the tobacco leaf. The function of JcMYB2 in salt and freezing tolerance were detected in transgenic plants.
Results
A genome-wide analysis identified 128 MYB genes, including 123 R2R3-MYBs, 4 R1R2R3-MYBs and 1 4R-MYB. All of the R2R3-MYBs are further classified into 19 groups which indicated functional conservation among previously identified groups of R2R3-MYB proteins. Among of these newly identified MYBs, the JcMYB2 belongs to group G11 and its expression is induced obviously by cold, salt and MeJA (Methyl Jasmonate) and slightly by ABA (abscisic acid). JcMYB2 is localized to the nucleus and has transcriptional activity. JcMYB2 overexpressing plants are more tolerant to salt and cold stress than wild type plants. Tissue specific expression profiles showed that the JcMYB2 gene was expressed ubiquitously throughout the plant, with higher expression levels observed in the root.
Conclusion
A comprehensive genome-wide analysis and phylogenetic relationship of R2R3-MYB subfamily in J. curcas present the global identification and functional prediction of JcR2R3-MYBs. Additionally, JcMYB2 regulates the stress response signaling networks by interacting with MeJA and ABA signaling pathway and functions in the root development of J. curcas.
Similar content being viewed by others
Background
J. curcas, a deciduous tree species belonging to the Euphorbiaceae family, is a drought resistant oil species widely distributed in tropical and subtropical areas, especially in Central and South America, Africa, India and Southeast Asia [1]. Among the potential biofuel crops, J. curcas has been gaining importance as the most promising oilseed, as it does not compete with the edible oil supplies. However, the lack of adequate genetic variation and non-availability of improved varieties limits its prospects of being a successful energy crop [2]. In addition, J. curcas plants are endowed with a high tolerance to conditions of drought and heat, moderate tolerance to salinity and heavy metals, but very low tolerance to low temperatures [3] which is the bottleneck for cultivation and commercialization of J. curcas. Low temperatures have persistent and detrimental effects on J. curcas crop establishment via the depletion of photosynthesis brought by chilling and freezing injuries [4]. The damage of cold stress may cause a sharp decrease in chlorophyll contents and membrane unsaturated fatty acids of J. curcas and even survival of seedlings [1].
Gao et al. suggest that besides the ROS (Reactive oxygen species) scavenging system, flavonoids from the phenylpropanoid pathway can also protect membrane lipids during cold stress in J. curcas seedlings exposed to temperatures as low as 4 °C [5]. A global view of the J. curcas transcriptome in response to cold stress revealed that 4,185 transcripts are possibly associated with cold resistance [6]. In our previous study, we found that expression of the JcERF gene is rapidly induced upon salinity, drought, ethylene and mechanical wounding treatments. Overexpression of JcERF in transgenic Arabidopsis enhances the salt and freezing tolerance [7]. Additionally, expression of JcDREB is induced by cold, salt and drought stresses, but not by ABA. Transgenic Arabidopsis plants over-expressing JcDREB exhibit enhanced tolerance to salt and freezing stresses [8]. Although we have made some progress in understanding the molecular mechanisms of J. curcas cold stress responses, the roles of transcriptional factors, especially for the MYB family which play a conserved role in the regulation of stress responses, has not been well illustrated.
According to the number of adjacent MYB repeats (R), MYB proteins can be classified into four subfamilies: MYB proteins with one repeat are named 1R-MYB (or MYB-related proteins); with two, 2R-MYB (R2R3-MYB); with three, 3R-MYB (R1R2R3-MYB); and with four, 4R-MYB (R0R1R2R3-MYB) [9, 10]. In plants, R2R3-MYB is the largest subfamily. The phylogenetic comparison of this superfamily in Arabidopsis and rice indicated that the Arabidopsis MYB superfamily undergo a rapid expansion after its divergence from monocots but before its divergence from other dicots [11]. Currently, there is some controversy about the evolution and categorization of subgroups. For instance, in Arabidopsis, 126 R2R3-MYB have been listed into 25 subgroups by Dubos et al. [10], which is different from Kranz et al., who categorized these MYBs into 22 subgroups [12]. Nevertheless, there are similar functions between members of each subgroup and there is cross-interaction among of them [12]. Recently, the increasing availability of plant genome sequences has facilitated a better understanding of this large gene family. In addition to Arabidopsis, genome wide characterization of the MYB family, especially for R2R3-MYB, have been completed in rice [13], maize [14], barely [15], foxtail millet [16], soybean [17], orange [18] and apple [19]. Comparative expression profile analysis of R2R3-MYB genes in these species suggested that MYB proteins play conserved and various roles in development, growth and regulation of the metabolism of plants [10]. MYB proteins have important roles in phytohormone signal transduction and various stress response pathways [20, 21]. The expression of AtMYB2 is up-regulated by ABA and the overexpression AtMYB2 enhances the drought tolerance of transgenic plants [22]. Under these conditions, the RD22 and AtADH1 are also up-regulated, which suggests that AtMYB2 functions in ABA-inducible gene expression under drought stress in plants [22, 23]. By interacting with ICE1, AtMYB15 acts as a negative regulator of freezing tolerance by suppressing the expression of CBF [24]. GENEVESTIGATOR analysis show that 44.67 and 47.21 % MYB genes in Arabidopsis are up- and down-regulated in cold stress, respectively [13]. It is thus reasonable that MYB proteins may be key regulatory nodes of plant responses to cold stresses. Therefore, in this study, we performed genome-wide identification of the MYB family in J. curcas, and further determined the function of the cold and salt responsive MYB2 gene in J. curcas.
Results
Identification of J. Curcas MYB genes
We used three approaches to identify the MYB protein encoding genes in J. curcas genome. To confirm the MYB protein models that were identified, all of the genes derived from the selected JcMYB candidate genes were examined using the online databases Pfam and SMART with default cutoff parameters. As a result, 123 typical non-redundant R2R3 MYB transcription factors, 4 R1R2R3 MYBs, and 1 4R-like MYB s were confirmed from the original data. This number is similar with that found in Arabidopsis. Additionally, the protein character of these 123 R2R3 MYB transcription factors were list in Additional file 1.
Phylogenetic analysis and functional prediction of J. Curcas R2R3-MYB
To evaluate the evolutionary relationships within the R2R3-MYB gene family, we performed a combined phylogenetic analysis of Arabidopsis and J. curcas MYB proteins using the neighbor joining method. Based on conserved DNA binding domain similarity and topology of their encoded proteins, we subdivided the 123 typical members of the J. curcas R2R3-MYB gene subfamily into 19 groups (designated G1-G19) according to the alignment results (Fig. 1). The alignment result was provided in Additional file 2. Generally, the number of orthologs were similar between Arabidopsis and J. curcas. However, some homologs were clustered remarkably by species within the same clade or the number of homologs from two species was greatly asymmetrical in one group or clade. For example, two AtMYBs and 17 JcMYBs were included in group G12.
Evolutionary relationships of JcR2R3-MYB. The evolutionary history was inferred using the Neighbor-Joining method. The statistical method is Maximum Likelihood (ML). The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the MYBs analyzed. The evolutionary distances were computed using the p-distance method and are in the units of the number of amino acid differences per site. The analysis involved 249 amino acid sequences. Evolutionary analyses were conducted in MEGA5.2. Among of these, JcMYB2 is a member of group 11 and identified by the red square
The functions of some Arabidopsis R2R3 MYB members have been well characterized experimentally, and phylogenetic analysis has identified several functional groups [18, 25, 26]. Most of JcR2R3-MYB proteins were clustered into the functional groups found in Arabidopsis (Fig. 1), including stress response, cell cycle, cell wall thickening, meristem formation, and secondary metabolism.
Cloning and expression profile of the JcMYB2 gene
The full length of JcMYB2 gene (GenBank accession: GU937787) is 908 bp and contains an ORF (open reading frame) of 570 bp that encodes a protein of 189 amino acids with a predicted molecular mass of 21. 9 kDa and a pI of 9.435. Analysis of the deduced amino acid sequence indicated that it is an R2R3-MYB protein with two imperfect repeat sequences in its MYB domain. Based on the phylogenetic tree of J. curcas and Arabidopsis MYB proteins, JcMYB2 clustered in group G11 with Arabidopsis AtMYB66, AtMYB23 and AtMYB0 (Fig. 1). There are two introns in the ORF of JcMYB2, with lengths of 73 and 72 bp, respectively. The gene structure of JcMYB2 is shown in Fig. 2a.
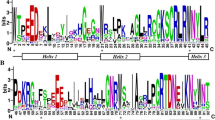
The gene structure and the dimensional structure of JcMYB2 protein. a The structure of JcMYB2 gene. There were two introns and three exons in this gene. The length of exon 1 and 3 included the length of the 5′ and 3′ UTR, respectively. b The alignment of JcMYB2 and its ortholog proteins of Arabidopsis. The red asterisk show the variant amino acid residues. R2 and R3 HTH represent the MYB repeat domain
The three-dimensional spatial structure of the JcMYB2 protein, predicted via CPHmodels-3.0, showed that JcMYB2 spatial structure has typical structure for R2R3-MYB proteins, namely two consecutive HTH (Helix-Turn-Helix) domains (Fig. 2b and Additional file 3); these constitute its DNA-binding domain and are predict to bind the major groove of DNA.
To determine whether JcMYB2 functions in stress responses in J. curcas, we assayed its expression following exposure of the plants to cold temperatures; high salt conditions; and both ABA and MeJA. Results showed that JcMYB2 transcripts were induced after 3 h cold stress treatment and reached their maximum at 6 h (Fig. 3a). Its expression pattern under salt stress was similar with that of cold (Fig. 3b). For ABA treatment, expression of JcMYB2 was initially induced after 1 h and accumulated continuously for 24 h (Fig. 3c). Additionally, under MeJA treatment JcMYB2 transcript was gradually increased and also reached its maximum at 6 h under MeJA treatment, then decreased sharply and returned to original levels (Fig. 3d). Under normal conditions, the expression of JcMYB2 in the root was much higher than that in the stem, leaf and cotyledon (Fig. 3e) which suggested that JcMYB2 was differentially expressed within different tissues of the plant.
Expression patterns of JcMYB2. a Expression pattern of JcMYB2 under 4 °C cultivate condition. b Expression pattern of JcMYB2 under 200 mM NaCl treatment. c Expression pattern of JcMYB2 under 100 μM ABA treatment. d Expression pattern of JcMYB2 under 100 μM MeJA treatment. e The organ specific expression pattern of the JcMYB2 gene. Transcript levels were determined by quantitative RT-PCR. Expression of J. curcas actin was used as an internal control. Error bars indicate standard deviation of three independent biological replications
Promoter isolation and activity analysis of JcMYB2
The promoter sequence of JcMYB2 was cloned using Tail-PCR and was subsequently confirmed by PCR designed according to the J. curcas genome database (Fig. 4). The structure of the JcMYB2 promoter region was analyzed by searching the plant cis-acting element database PLACE (Plant Cis-Acting regulatory DNA Elements) and Plant CARE (Plant cis-acting regulatory element database); the results are shown in Table 1. This promoter contains total putative 72 cis-elements classified in to 11 types, including those involved in ABA responses and stress tolerance, low temperature stress tolerance, JA-induced plant disease resistance, SA-induced pathogen resistance and meristem development.
GUS staining of transgenic Arabidopsis harboring Pro-JcMYB2::GUS revealed that the JcMYB2 promoter is very active in the root, but has only slightly activity in the leaf and stem (Fig. 5b). Cold, salt, ABA and MeJA could induce GUS expression in transgenic Arabidopsis (Fig. 5c-f).
The promoter activity of JcMYB2. Detection of JcMYB2 promoter activity by GUS staining. a wild type. b the transgenic plant under normal cultural condition. c seedlings under 4 °C cultivate condition for 6 h. d seedlings under salt treatment (treated with 200 mM NaCl for 6 h). e seedlings under ABA treatment (treated with 100 μM ABA for 3 h). f seedlings under MeJA treatment (treated with 100 μM MeJA for 6 h). B, C, D, E and F show GUS staining signal except for A. The GUS staining analysis of transgenic Arabidopsis harboring Pro-JcMYB2:: GUS revealed that the JcMYB2 promoter was active in transgenic seedlings. C, D, E and F display GUS staining signal which proved that its activity of JcMYB2 promoter will be increased under cold, salt, ABA and MeJA treatment
Transcriptional activity and subcellular localization of the JcMYB2 protein
The transcriptional activity of JcMYB2 was tested using a yeast one-hybrid assay. The yeast strain AH109 containing the vector pBridge (as a negative control) could not grow on SD medium without His and Trp (SD/-His-Trp). However, cells harboring the pBridge-JcMYB2 and pBridge-JcERF (as a positive control) could grow normally on the same medium and exhibited blue in β-galactosidase assay (Fig. 6a-c). These results indicate that JcMYB2 can function as a transcriptional activator.
Transactivational assay of JcMYB2. The full-length ORF of JcMYB2 was fused with pBridge, and the transformed AH109 yeasts were selected from SD-Trp-His medium. JcERF was used as a positive control, and empty pBridge vector was used as a negative control. a The position of each transformed yeast cell. b The growth status of the transformed AH109 cells on the SD medium. c the reporter activity detected by β-galactosidase activity assay
To determine the subcellular localization of JcMYB2, we visualized a JcMYB2-GFP fusion protein in tobacco epidermal cells using confocal microscopy. In these cells, GFP signal was observed exclusively in the nucleus, while GFP fluorescence in control, pCAMBIA1302-GFP was distributed in the whole cell (Fig. 7). Thus, the result suggested that JcMYB2 is a nuclear protein.
Subcellular localization of JcMYB2 protein. Transient expression of pCAMBIA1302- JcMYB2 fusion and pCAMBIA1302 construct in tobacco epidermal cells. Green fluorescence was observed using a confocal microscope at 48 h after A. tumefaciens infiltration. Micrographs showing cells expressing GFP (control, upper lane) or JcMYB2::GFP (bottom lane) fusion protein. From left to right, the pictures showed DAPI, fluorescent-field illumination, bright-field, and overlay of three illuminations. Bar = 30 μm
Heterologous expression of JcMYB2 enhances salt and freezing tolerance of Arabidopsis
To investigate the biological function of JcMYB2, ten day old seedlings of transgenic and wild type Arabidopsis were transferred to MS medium supplemented with 200 mM NaCl after germination on MS medium. The survival percentage of JcMYB2 transgenic Arabidopsis was more than 85.6 % under salt stress, which was significantly higher than that of wild type Arabidopsis (lower than 14.8 %) (Fig. 8). For the freezing tolerance test, three-week-old plants were exposed to −8 °C for 20 h, and then recovered in chambers at 23 °C for 7 d. The transgenic plants expressing JcMYB2 were able to grow after being removed from sub-zero temperatures, while the wild type plants withered and died under these conditions (Fig. 8). Taken together, these results show that overexpression of JcMYB2 in Arabidopsis confers tolerance to both high salt and low temperature stresses.
Cold and salt stress response of Arabidopsis transgenic plants overexpressing JcMYB2. Three-week-old plants, including wild type and transgenic plants, were exposed to −8 °C for 20 h, after which the temperature was returned to 23 °C for 7 days. The growth of Wild type and transgenic lines plants were grown on MS media for 10 days and then transferred onto the MS with 200 mM NaCl for salt stress. When the difference of phenotype between Wild type and transgenic lines were apparent, the photograph were taken
Discussion
Identification and functional prediction of the J. curcas MYB family
To identify the MYB transcription factor-encoding genes in J. curcas, we searched the entire J. curcas genome and two databases for genes that encode proteins containing the MYB DNA binding domain. As is well known, the MYB family is the most abundant transcription factor family in plants and R2R3-MYB is the largest subfamily. Correspondingly, there are 157 R2R3-MYB proteins in the maize [14], 100 in the sweet orange [18], 126 in Arabidopsis, 109 in rice [13], 252 in soybean [17] and 192 in Populus [27]. Additionally, there are 222 typical R2R3-MYB proteins, five R1R2R3-MYB proteins, and two 4R-like MYB proteins in the apple genome [19]. While the number of R2R3-MYB-encoding genes has expanded in different species, the number of R1R2R3-MYB and 4R-MYB genes has not changed. All vertebrate MYB proteins are in the 3R-MYB subfamily, which is also present in almost every plant studied [28], suggesting that the 3R-MYB encoding genes represent an ancient and evolutionarily conserved family. In both plants and vertebrates, 3R-MYB proteins regulate progress through cell cycle transitions [29].
These J. curcas R2R3-MYB proteins were divided into 19 groups, and the phylogenetic results was consistent with the recent reports [18, 26], which confirmed that our results placed the J. curcas proteins into the correct subfamilies. In general, the functions of members of the same clade seem highly but not absolutely conserved across different plants. The functions of some Arabidopsis R2R3-MYB proteins have been well characterized experimentally, and phylogenetic analysis has identified some functional groups [18, 25, 26]. Therefore, it is useful to identify the orthologs between plants based on their evolutionary relationships. Although the functions of most JcMYBs have yet to be characterized, all of the JcMYB proteins clustered into Arabidopsis functional groups (Fig. 1) which could provide useful information for predicting and studying the functions of JcMYBs within each clade. Many JcR2R3-MYBs were grouped into G13, G15, G17 and G18, whose members have been shown to be involved in stress response in Arabidopsis (Fig. 1), which thus facilitated identifying the JcMYBs that may play roles in the response to stress conditions.
JcMYB2 is a typical R2R3-MYB transcription factor
We isolated one R2R3-JcMYB2 gene from J. curcas by RACE PCR. The targeting experiment confirmed that JcMYB2 localizes to the nucleus (Fig. 7) and a yeast one hybrid assay suggested that JcMYB2 possesses transcriptional activation ability (Fig. 6). In addition, two introns of JcMYB2 are located up- and down-stream of the sequence encoding the conserved R2R3 repeat domain (Fig. 2a), which was similar to its orthologous in Arabidopsis. Thus, we inferred that JcMYB2 might originate from the same ancestral gene as Jcr4S12250.20, AT3G27920 (AtMYB0) and AT5G14750 (AtMYB66), and some variation occurred in the DNA binding domain, though the length and position of the introns remain highly conserved.
Besides, JcMYB2 was clustered in the G11 with other MYB proteins from Arabidopsis (Fig. 2b), including AtMYB0, AtMYB23 and AtMYB66. The first helix of the second repeat domain is essential for the formation of the ternary complex [30, 31]. For AtMYB0 and AtMYB66, some variation of these residues, especially the mutation of the K residues will prevent their interaction with other proteins. According to the alignment of the full length protein of these four MYBs, in addition to the highly conserved amino acid residues, we observed that K is replaced by N residue in JcMYB2 (Fig. 2b). Additionally, there is another conserved residue Q is replaced by E in JcMYB2. Therefore, compared with AtMYB0 and AtMYB66, we hypothesized that JcMYB2 might play a different role in J. curcas.
JcMYB2 is involved in growth regulation, development and tress tolerance
MYB proteins have been reported to be involved in response to cold stress in rice. For instance, overexpression of OsMYB4 significantly enhanced tolerance to chilling and freezing stress in transgenic Arabidopsis [32]. Ma et al. reported that OsMYB3R-2 participates in the cold signaling pathway by targeting the cell cycle and a putative DREB/CBF transcription factor [33]. Moreover, a recent study revealed that an R2R3-MYB gene, OsMYB2, is induced by salt, cold, and dehydration stress. OsMYB2 overexpressing plants are more tolerant to salt, cold and more sensitive to abscisic acid than wild type plants [34]. Expression pattern analysis of the 156 MYB genes identified in soybeans suggests that expression of 43 of these genes change upon treatment with cold, salt and/or drought stress [35]. GmMYBJ1 displays similarities to the typical R2R3MYB proteins reported in other plants and overexpressing GmMYBJ1 causes enhanced tolerance to drought and cold stresses, which indicates that GmMYBJ1 has the potential to be utilized in transgenic breeding lines to improve abiotic stress tolerance [36]. PtsrMYB isolated from the trifoliate orange shares the highest degree of identity with AtMYB109 and is up-regulated by abiotic stresses such as dehydration, salt, cold and ABA treatment [37].
The expression of JcMYB2 was significantly enhanced under salt and cold stress, as well as induced by ABA and MeJA treatment. Therefore, we inferred that JcMYB2 is involved in ABA-dependent cold and salt tolerance and JA-mediated disease resistance signaling pathways. There is significant cross-talk among abiotic stress response mediated by ABA and the biotic stress response mediated by JA. Arabidopsis mutants with ABA insensitivity have the strong resistance against pathogens. On the other hand, excess exogenous ABA results in wild type plant susceptibility to the pathogens [38]. However, the infection will leads to an increase in the endogenous ABA [39]. Recent studies have reported that many kinds of transcription factors are induced expression by ABA and JA [40–45]. For instance, there is a report that the expression of AtMYB2 and AtMYC2 increases under ABA treatment, and their over-expression enhances the drought and salt tolerance [22]. Further studies have revealed that these genes also play important roles in the JA-mediated pathogen resistance signaling pathway, and AtMYC2 especially has been considered the essential junction between biotic and abiotic stress [46, 47].
In plants, gene expression is determined by the promoter. An analysis of the JcMYB2 promoter revealed several stress response elements, including those involved in low temperature stress tolerance, ABA responses and stress tolerance, JA-induced plant disease resistance and SA induced pathogen resistance. The expression pattern of JcMYB2 and GUS staining analysis validated the potential function of these elements. It is a remarkable that 10 MYB binding sites present in the promoter JcMYB2, which implies that JcMYB2 might be regulated by itself or other MYB proteins. Additionally, the transgenic expression of JcMYB2 in Arabidopsis enhanced salt and cold tolerance in our study. Thus we inferred that JcMYB2 functions in the ABA-dependent and MeJA-mediated abiotic and biotic stress responses.
In addition, many growth and development related elements were also found in the JcMYB2 promoter, such as light response elements, meristem development, endosperm development and late pollen development. In this study, the JcMYB2 showed clear differential expression with high expression in the root and low expression in the stem, leaf and cotyledon. Interestingly, the AtMYB66, the Arabidopsis ortholog of JcMYB2, is highly expressed in the root and hypocotyl, and only lowly expressed in other organs of Arabidopsis seedlings. AtMYB66 regulates differentiation of the non-root hair cell in the root and the non-stomata cell in the hypocotyl [48, 49], AtMYB0, another ortholog of JcMYB2, also controls the differentiation of the root hair and the stem hair [50]. There is some similarity in the expression of JcMYB2 and that of AtMYB0 and AtMYB66, which implies that JcMYB2 might have a similar function in the control of root cell differentiation, which need further investigation. Taken together, these results suggest that JcMYB2 is not only involved in cold and salt stress response, but also functions in MeJA-mediated biotic stress, and regulation of root development and growth. To some extent, these results also showed the feasibility of predicting protein functions through phylogenetic analysis.
Conclusion
In conclusion, this study presents a genome-wide identification of the MYB gene family in J. curcas. A total of 123 R2R3-MYB proteins were phylogenetically classified into 19 distinct groups and putative function of R2R3-MYB proteins was assigned based on phylogenetic results. Additionally, JcMYB2, located in the nucleus and activated the downstream gene expression, could enhance the salt and cold resistance of the transgenic plants. The results presented here will be helpful for future studies of the biological functions of JcMYB proteins.
Methods
Identification and sequence analysis of MYB proteins in J. curcas
Three approaches were used to identify the members of the JcMYB gene family in J. curcas. First, a HMM profile of the MYB DNA-binding domain (PF00249) from the Pfam database (http://pfam.sanger.ac.uk/) was used for the identification of MYB genes in J. curcas genome (http://www.kazusa.or.jp/jatropha/). The default parameters were employed; the number of search results was set as 200. Second, we used MYB and J. curcas as keywords to search the NCBI (http://www.ncbi.nlm.nih.gov/) and all of the retrieved sequences were download. Third, the protein sequence and their DNA binding domain of MYB proteins were download from the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/). All of the protein sequences deriving from the selected MYB candidate genes were examined with the domain analysis programs of Pfam and SMART (http://smart.embl-heidelberg.de/) with default cutoff parameters. Finally, all of the protein sequences were compared with known MYB sequences by alignment to verify that the candidate sequences from J. curcas encoded MYB-containing proteins.
Phylogenetic analysis and function prediction of the JcR2R3-MYBs
The Arabidopsis R2R3-MYB proteins and their conserved DNA binding domains were downloaded from the Plant Transcription Factor Database. The conserved DNA binding domain of R2R3-MYBs from J. curcas and Arabidopsis were aligned by MUSCLE in MEGA 5.2 and adjusted manually. The default parameters for gap open and gap extend is −2.9 and 0, respectively. Then, the phylogenetic trees were constructed based on the multiple sequence alignment by using the neighbor-joining method in MEGA 5.2 with 1000 bootstrap replicates. The images of the phylogenetic trees were drawn in MEGA5.2. Additionally, the biological functions of some JcMYBs were predicted based on the aforementioned phylogenetic tree according to orthology. In addition, the three-dimensional spatial structure of the JcMYB2 protein was predicted via CPHmodels-3.0.
Plant materials and treatments
The seeds of J. curcas were grown in pots with 1:1(v/v) vermiculite vs peat medium and incubated at 28 °C with a 16 h light/8 h dark photoperiod for three weeks. For cold treatment, the seedlings were transferred into a 4 °C growth chamber. For salt and ABA treatment, the seedlings were washed carefully and transferred into the solutions of 200 mM NaCl and 100 μM ABA, respectively. For MeJA treatment, 100 μM solution was sprayed onto the surface of the seedlings. Both the control and stress treated seedlings were harvested at various periods (1, 3, 6, 12 and 24 h), flash frozen in liquid nitrogen and stored at −80 °C for further analysis. The root, stem, leaf and cotyledon were collected for tissue expression analysis.
Isolation of the JcMYB2 gene and promoter
Total RNA was extracted from J. curcas samples as described by Tang [8] and digested by RNase-free DNase I (TaKaRa, Japan) to remove the genomic DNA. The genomic DNA of J. curcas was extracted using a DNAsecure PlantKit (Tiangen Biotech, Beijing). The concentrations of DNA and total RNA were determined using a Nano Drop cDNA for amplification of the 5′ and 3′ ends of JcMYB2 was prepared using a SMARTTM RACE cDNA Amplification Kit (Clontech, Japan). The RACE primers of JcMYB2 were designed based on the results of Sanger sequencing. The 5′ and 3′ end of JcMYB2 were cloned following the manufacturer’s instructions. The reaction conditions were as follows: 94 °C for 4 min; 36 cycles of 94 °C for 30 s, 68 °C for 30 s, 72 °C for 50 s; followed by 72 °C for 10 min. The RACE results was provided in Additional file 4. Full-length JcMYB2 cDNA was amplified using cDNA synthesized for the 3′ RACE as a template. The genomic sequences of JcMYB2 were cloned using the same primers. The promoter of JcMYB2 was cloned by using the TAIL PCR (thermal asymmetric interlaced PCR) with a Genome Walking Kit (TaKaRa, Japan). All the PCR products were cloned into the pMD19-Tvector (TaKaRa, Japan) and sequenced by Sangon Biotech (Shanghai).
To determine the activity of the JcMYB2 promoter, the CaMV 35S promoter of pCAMBIA1301 was replaced by the promoter of JcMYB2 to promote the expression of GUS. The recombinant plasmid was transformed into Agrobacterium tumefaciens EHA105, which was used to infect Arabidopsis. GUS histochemical staining was used to test the activity of the JcMYB2 promoter. All of the primers were list in Additional file 5.
Expression patterns of JcMYB2
Real-time quantitative PCR (qPCR) was completed on a qPCR machine (MX3000p, Strategene, CA, USA) using the SYBR PrimeScript RT-PCR Kit (TaKaRa), and the reaction condition was following: program of 40 cycles (95 °C for 5 s and 60 °C for 20 s). The 2−ΔΔCT method was used to analyze the data and expression of J. curcas actin was monitored as an internal control. Information about the other primers used in qPCR can be found in Additional file 5.
Transcriptional activity investigation
The entire coding region of JcMYB2 was inserted into the BamHI and SalI sites of pBridge to yield a fusion protein in frame with the GAL4 DNA binding domain. The primers used for PCR are listed in Additional file 6. The recombinant plasmid was sequenced to ensure that it could express proteins accurately. The recombinant plasmid was transformed into yeast AH109 with the reporter genes His3 and LacZ according to the manufacturer’s instructions (Clontech, Palo Alto CA, USA). pBridge-JcERF [8] was used as a positive control. The empty pBridge vector served as a negative control. Transformed yeasts were cultured on SD medium without His and Trp. Subsequently, in order to test the expression of the LacZ gene, a colony lift filter assay was performed using o-nitrophenyl-β-D-galactopyranoside (ONPG) as a substrate to determine whether the JcMYB2 protein possesses transcriptional activity.
Subcellular localization of JcMYB2 protein
The subcellular localization of JcMYB2 protein was determined as described by Peng [44]. The entire coding region of the target gene without a stop codon was amplified by PCR and inserted into the NcoI and SpeI sites of the vector pCAMBIA1302, which fuse GFP, in frame, to the 3′ end of JcMYB2. pCAMBIA1302 -JcMYB2-GFP construct was introduced into Agrobacterium tumefaciens strain EHA105. The tobacco leaf was infiltrated by the Agrobacterium to express the GFP fusion protein. Following incubation in growth chambers for 48 h, and then the injected leaf was stained by 100 ng/ml DAPI (4′–6-diamidino-2-phenylindole). After that, DAPI and GFP fluorescence in tobacco leaves was imaged by confocal scanning microscope (Leica, Göttingen, Germany).
Analysis of salt and freezing tolerance in transgenic plants
The full-length JcMYB2 cDNA was ligated into p3301-121 (reconstructed by our Lab) under the control of the CaMV 35S promoter. Empty vector was used as a control. The recombinant plasmid was transformed into Agrobacterium tumefaciens EHA105. Arabidopsis were transformed by the floral dip method. The T0 seeds were harvested, dried at 25 °C, and plated on half-strength sterile MS medium with 10 mg/l PPT selection pressure; subsequently surviving T1 seedlings were transferred to soil to set T2 seeds. T2 seeds and seedlings were used for the following analysis of salt and freezing tolerance.
Transgenic plants were grown in 8 cm pots filled with a mixture of peat/vermiculite (1:1, v/v) under light at 23 °C for 4 weeks. Three-week-old plants were exposed to freezing stress. Freezing stress was conducted by exposure of plants to a temperature of −8 °C for 20 h, after which the temperature was returned to 23 °C for 7 days. Transgenic plants were grown on MS media for 10 days and then transferred onto the MS with 200 mM NaCl for culture.
References
Liang Y, Chen H, Tang MJ, Yang PF, Shen SH. Responses of Jatropha curcas seedlings to cold stress: photosynthesis-related proteins and chlorophyll fluorescence characteristics. Physiol Plant. 2007;131(3):508–17.
Johnson TS, Eswaran N, Sujatha M. Molecular approaches to improvement of Jatropha curcas Linn. as a sustainable energy crop. Plant Cell Rep. 2011;30(9):1573–91.
Garcia-Almodovar C, Gimeno V, Nieves M, Diaz-Lopez L, Simon I, Garcia-Sanchez F. Improving the tolerance of Jatropha curcas L. plants to abiotic stresses. CAB Reviews. 2014;9(004):1–10.
Ploschuk E, Bado L, Salinas M, Wassner D, Windauer L, Insausti P. Photosynthesis and fluorescence responses of Jatropha curcas to chilling and freezing stress during early vegetative stages. Environ Exp Bot. 2014;102:18–26.
Gao J, Jiang N, Qin X, Zhu X, Ai T, Peng T, Wu J, Xu Y, Chen F. Physiological and metabolic responses of Jatropha to chilling stress. Int J Agric Biol. 2013;15(5):871–7.
Wang H, Zou Z, Wang S, Gong M. Global analysis of transcriptome responses and gene expression profiles to cold stress of Jatropha curcas L. PLoS One. 2013;8(12):e82817.
Tang M, Sun J, Liu Y, Chen F, Shen S. Isolation and functional characterization of the JcERF gene, a putative AP2/EREBP domain-containing transcription factor, in the woody oil plant Jatropha curcas. Plant Mol Biol. 2007;63(3):419–28.
Tang M, Liu X, Deng H, Shen S. Over-expression of JcDREB, a putative AP2/EREBP domain-containing transcription factor gene in woody biodiesel plant Jatropha curcas, enhances salt and freezing tolerance in transgenic Arabidopsis thaliana. Plant Sci. 2011;181(6):623–31.
Du H, Wang Y, Xie Y, Liang Z, Jiang S, Zhang S, Huang Y, Tang Y. Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 2013;20(5):437–48.
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–81.
Chen Y, Yang X, He K, Liu M, Li J, Gao Z, Lin Z, Zhang Y, Wang X, Qiu X. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60(1):107–24.
Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. The Plant Journal. 1998;16(2):263–76.
Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics. 2012;13(1):544.
Du H, Feng B, Yang S, Huang Y, Tang Y. The R2R3-MYB transcription factor gene family in maize. PLoS One. 2012;7(6):e37463.
Tombuloglu H, Kekec G, Sakcali MS, Unver T. Transcriptome-wide identification of R2R3-MYB transcription factors in barley with their boron responsive expression analysis. Mol Gen Genomics. 2013;288(3–4):141–55.
Muthamilarasan M, Khandelwal R, Yadav CB, Bonthala VS, Khan Y, Prasad M. Identification and molecular characterization of MYB transcription factor superfamily in C4 model plant foxtail millet (Setaria italica L.). PLoS One. 2014;9(10):e109920.
Du H, Yang S, Liang Z, Feng B, Liu L, Huang Y, Tang Y. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC plant biology. 2012;12(1):106.
Liu C, Wang X, Xu Y, Deng X, Xu Q. Genome-wide analysis of the R2R3-MYB transcription factor gene family in sweet orange (Citrus sinensis). Mol Biol Rep. 2014;41(10):6769–85.
Cao Z, Zhang S, Wang R, Zhang R, Hao Y. Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS One. 2013;8(7):e69955.
Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants. 2013;19(3):307–21.
Li C, Ng CK, Fan L. MYB transcription factors, active players in abiotic stress signaling. Environ Exp Bot. 2014.
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. The Plant Cell. 2003;15(1):63–78.
Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought-and abscisic acid-regulated gene expression. The Plant Cell. 1997;9(10):1859–68.
Agarwal M, Hao Y, Kapoor A, Dong C, Fujii H, Zheng X, Zhu J. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006;281(49):37636–45.
Matus JT, Aquea F, Arce-Johnson P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biology. 2008;8(1):83.
Chen N, Yang Q, Pan L, Chi X, Chen M, Hu D, Yang Z, Wang T, Wang M, Yu S. Identification of 30 MYB transcription factor genes and analysis of their expression during abiotic stress in peanut (Arachis hypogaea L.). Gene. 2014;533(1):332–45.
Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009;149(2):981–93.
Haga N, Kato K, Murase M, Araki S, Kubo M, Demura T, Suzuki K, Müller I, Voß U, Jürgens G. R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development. 2007;134(6):1101–10.
Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A. G2/M-phase–specific transcription during the plant cell cycle is mediated by c-Myb–like transcription factors. The Plant Cell. 2001;13(8):1891–905.
Schaart JG, Dubos C, Romero De La Fuente I, Houwelingen AM, Vos RC, Jonker HH, Xu W, Routaboul JM, Lepiniec L, Bovy AG. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 2013;197(2):454–67.
Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL. Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc Natl Acad Sci. 2000;97(25):13579–84.
Pasquali G, Biricolti S, Locatelli F, Baldoni E, Mattana M. Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep. 2008;27(10):1677–86.
Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009;150(1):244–56.
Yang A, Dai X, Zhang W. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot. 2012;63:err431.
Liao Y, Zou H, Wang H, Zhang W, Ma B, Zhang J, Chen S. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 2008;18(10):1047–60.
Su L, Li J, Liu D, Zhai Y, Zhang H, Li X, Zhang Q, Wang Y, Wang Q. A novel MYB transcription factor, GmMYBJ1, from soybean confers drought and cold tolerance in Arabidopsis thaliana. Gene. 2014;538(1):46–55.
Sun P, Zhu X, Huang X, Liu J-H. Overexpression of a stress-responsive MYB transcription factor of Poncirus trifoliata confers enhanced dehydration tolerance and increases polyamine biosynthesis. Plant Physiol Biochem. 2014;78:71–9.
Ryerson E, Li A, Young J, Heath M. Changes in abscisic acid levels in bean leaves during the initial stages of host and nonhost reactions to rust fungi. Physiol Mol Plant Pathol. 1993;43(4):265–73.
Audenaert K, De Meyer GB, Höfte MM. Abscisic acid determines basal susceptibility of tomato toBotrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 2002;128(2):491–501.
Puranik S, Sahu PP, Srivastava PS, Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17(6):369–81.
Tripathi P, Rabara RC, Rushton PJ. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta. 2013;239:1–12.
Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15(5):247–58.
Berri S, Abbruscato P, Faivre-Rampant O, Brasileiro A, Fumasoni I, Satoh K, Kikuchi S, Mizzi L, Morandini P, Pè M, Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC plant biology. 2009;9(1):120.
Peng X, Ma X, Fan W, Su M, Cheng L, Alam I, Lee BH, Qi D, Shen S, Liu G. Improved drought and salt tolerance of Arabidopsis thaliana by transgenic expression of a novel DREB gene from Leymus chinensis. Plant Cell Rep. 2011;30:1493–502.
Peng X, Zhang L, Zhang L, Liu Z, Cheng L, Yang Y, Shen S, Chen S, Liu G. The transcriptional factor LcDREB2 cooperates with LcSAMDC2 to contribute to salt tolerance in Leymus chinensis. Plant Cell Tissue Organ Cult. 2013;113:245–56.
Mengiste T, Chen X, Salmeron J, Dietrich R. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. The Plant Cell. 2003;15(11):2551–65.
Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. The Plant Cell. 2004;16(7):1938–50.
Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99(5):473–83.
Koshino-Kimura Y, Wada T, Tachibana T, Tsugeki R, Ishiguro S, Okada K. Regulation of CAPRICE transcription by MYB proteins for root epidermis differentiation in Arabidopsis. Plant Cell Physiol. 2005;46(6):817–26.
Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67(3):483–93.
Acknowledgement
This work was supported by the Knowledge Innovation Program through the Chinese Academy of Sciences (KZCX2-YW-359–2) and the National Natural Science Foundation of China (31270653), Huimin Technology Demonstration Project of the National Modern Agricultural Science and Technology Achievements City (Z151100001015008).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PXJ performed the genome-wide analysis of JcMYBs and wrote this manuscript. LH performed the plasmid construction and the yeast one hybrid. WD carried out the subcellular localization, promoter clone and qPCR experiments. SSH was responsible for the overall concept and experimental designs, data integration, analysis and interpretation, and manuscript preparation. All authors approved the final manuscript.
Additional files
Additional file 1:
The character of these 123 R2R3 MYB proteins. (XLSX 38 kb)
Additional file 2:
The alignment file of MYB binding domain of Arabidopsis and Jatropha curcas. Alignment analyses were conducted in MEGA5.2. (FAS 95 kb)
Additional file 3:
The predicted three dimensional structure of JcMYB2. Arrow: β-strand; Helix: α-helix; Single line: Turn. (TIF 800 kb)
Additional file 4:
The sequence of the RACE PCR. (XLSX 9 kb)
Additional file 5:
The composition of MYB proteins in plants. (XLSX 10 kb)
Additional file 6:
Primers used in this study. (XLSX 9 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Peng, X., Liu, H., Wang, D. et al. Genome-wide identification of the Jatropha curcas MYB family and functional analysis of the abiotic stress responsive gene JcMYB2 . BMC Genomics 17, 251 (2016). https://doi.org/10.1186/s12864-016-2576-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-016-2576-7