Abstract
A comparative metabolomic analysis was performed using purple potato tubers and derived red mutant tubers to investigate the regulation of anthocyanin biosynthesis. The metabolome analysis revealed 19 key differential metabolites including 10 flavonoids and 4 nucleotides. As a differential metabolite, pelargonidin was responsible for the mutant, and other metabolites had a correlation with it. To further analyse the regulatory mechanism of the differential metabolites, correlation analyses of metabolome and transcriptome were done. Among the differential metabolites, seven flavonoids and F3′5′H were in the flavonoid metabolic pathway. F3′5′H is responsible for the anthocyanin biosynthesis branch in potato tubers. To further analyse the regulatory mechanism of F3′5′H, the transcriptome analysis identified 63 differentially expressed genes, which highly correlated with F3'5'H. Furthermore, correlations among the key differential metabolites and the differentially expressed genes indicated that 11 differential genes and 10 differential metabolites presented 76 significant correlations. These 11 genes mainly encoded serine/threonine protein kinase, UMP-CMP kinase, MADS-box transcription factor, and WD repeat-containing protein. The 10 differential metabolites were involved in pelargonidin, luteolin, kaempferol-3-O-rutinoside, eriodictyol-7-O-glucoside, and uridine monophosphate. Through yeast one-hybrid experiments, the proteins interacting with the promoter of F3'5'H were screened: ethylene-responsive transcription factor ERF011, gibberellin-regulated protein 9-like, and serine-threonine protein kinase, transcription factor LHW bHLH. These results will provide valuable insights into the regulation mechanism of the anthocyanin biosynthesis branch in potatoes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato (Solanum tuberosum L.) is the fifth largest cultivated crop in the world and has been regarded as one of the staple food crops. In red and purple tubers of some potato cultivars, anthocyanins accumulate (Giusti et al. 2014; Brown et al. 2007; Han et al. 2006). Anthocyanins possess excellent antioxidant properties (Galvano et al. 2004; Prior 2004; Beattie et al. 2005) and thus can protect human beings from many chronic diseases, such as cancer, memory disorder, and diabetes (Long et al. 2018; Chen et al. 2019; Yan et al. 2016; Saito et al. 2016). Owing to the efficacy of anthocyanins and the important roles of the potatoes, studying anthocyanin biosynthesis of potato has a significant meaning for the improvement of public health.
Anthocyanins are flavonoid compounds and can be basically classified into six types by the numbers of hydroxyl groups and methyl groups, namely, pelargonidin, cyanidin, delphinidin, peonidin, petunidin, and malvidin (Martin and Gerats 1993). The relative contents and types of anthocyanin determine the final colour of fruits and other organs (Cutanda-Perez et al. 2009; Castellarin et al. 2006).
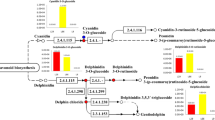
Anthocyanins are biosynthesised, starting from phenylalanine, by a so-called anthocyanin biosynthesis pathway. In this pathway, dihydrokaempferol involves in three branches of biosynthesis of anthocyanins, pelargonidin, cyanidin, and delphinidin (Holton and Cornish 1995; Bogs et al. 2006), which also biosynthesise their derivatives through glycosylation, methylation and acylation (Cavallini et al. 2014). The branches are controlled by flavonoid 3',5'-hydroxylase (F3'5'H) and flavonoid 3′-hydroxylase (F3'H) (Jeong et al. 2006), respectively. F3'5'H is responsible for biosynthesis of blue delphinidin-based anthocyanins (Lu and Yang 2006; Wessinger and Rausher 2015), while F3'H is responsible for biosynthesis of purple cyanin-based anthocyanins (Momose et al. 2013). Without F3'5'H and F3'H, red pelargonidin is biosynthesised (Holton and Cornish 1995; Liu et al. 2018). As the key enzyme controlling anthocyanins biosynthesis, F3′5′H catalyses the oxidation of two hydroxyls at the 3′ and 5′ sites of dihydroflavonol, and the reaction yields delphinidin-based anthocyanins (Olsen et al. 2010). Furthermore, the genes encoding the enzyme have been cloned in many species, such as potato, grape, and Dendrobium moniliforme (Momose et al. 2013; Whang et al. 2011; Falginella et al. 2010). Although the mechanism of action of F3′5′H and the genes encoding the enzyme have been studied, the genes and metabolites regulating the enzymes and the regulatory mechanism remain unknown.
With the development of omics technologies, anthocyanin biosynthesis has been intensively studied in many plants by combining analyses of metabolome, transcriptome, and proteome (Degu et al. 2014; Tohge et al. 2005; Wang et al. 2017a). In grapevine, the regulation of anthocyanin accumulation relied on the grape varieties through metabolic profile and transcript data (Degu et al. 2014). A comprehensive analysis of the metabolome and transcriptome in Arabidopsis thaliana identified several novel genes involved in anthocyanin biosynthesis (Tohge et al. 2005). The dynamics in transcriptome and metabolome of maize revealed the relationship between primary metabolism and anthocyanin accumulation (Hu et al. 2016). An analysis of metabolome and proteome showed anthocyanin accumulation dynamics in grape and fig (Wang et al. 2017a, b). Metabolites and transcriptional profiling analysis revealed the molecular regulation of anthocyanin balance in tea plants (Mei et al. 2021).
Previously, we extensively discussed the candidate genes of anthocyanin transformation through the comparative transcriptome analysis of a wild purple potato SD92 and its red mutant SD140 (Liu et al. 2018). SD140 was obtained through in vitro regeneration of SD92 tuber slices. In the current study, the differential metabolites between SD92 and SD140 were identified and their relationships were further explored to acquire the candidate metabolites regulating anthocyanin biosynthesis. Furthermore, the molecular mechanism of the anthocyanin biosynthesis branch regulated by F3′5′H in potatoes was studied by correlation analyses among the differential metabolites and differential genes. The results in this study will enrich the information about the anthocyanin metabolic pathway and will be beneficial to breeding anthocyanin-rich potato cultivars.
Materials and Methods
Plant Materials
The tetraploid SD92 with purple skin and purple flesh tuber and SD140, a red skin and flesh mutant identified from in vitro regenerated SD92 tuber slices, were used (Liu et al. 2018). SD92 and SD140 were planted in a greenhouse at the Institute of Vegetables and Flowers, Shandong Academy of Agricultural Sciences at the normal sowing time (August 10, 2016) in Jinan. Harvested fresh tubers (diameter 4–5 cm) were sliced into small pieces before being frozen in liquid nitrogen. Each material had 6 biological replicates, namely, SD92-1, SD92-2, SD92-3, SD92-4, SD92-5, SD92-6, SD140-1, SD140-2, SD140-3, SD140-4, SD140-5, SD140-6.
Extraction of Metabolites
The frozen tubers were ground to powder in liquid nitrogen with a mortar, and 100 mg of the powder was transferred to a 5-mL centrifuge tube, mixed with precooled (−20°C) 1000 μL methanol (Thermo, USA), vortexed for 30 s, and then ultrasonicated for 30 min. After ultrasonication, 750 μL chloroform (Sigma, USA), and 800 μL ddH2O (precooled at 4°C) were added to the tube which was then vortexed for 1 min. Mixture was centrifuged at 10000 rpm for 10 min at 4°C and 1000 μL supernatant was collected and then concentrated in a vacuum centrifugal concentrator for metabolome analysis.
Metabolite Profiling Analysis
The metabolite profiles of 12 samples were analysed via ultraperformance liquid chromatography (UPLC) (UHPLC LC-30A, Shimadzu, Japan) and triple quadrupole time-of-flight tandem mass spectrometry (Triple-TOF–MS/MS) (Triple TOFTM 5600 System, AB, America). Before these experiments, all sample extracts with four replicates were mixed for metabolite profiling analysis in the positive and negative ESI modes for quality control. Chromatographic separation was performed on an Acquity UPLC BEH C18 column (2.1 mm × 100 mm × 1.7 μm; Waters, America) by using 0.1% formic acid (TCI, Japan) in ultrapure water as mobile phase A and 0.1% formic acid in acetonitrile (Thermo, USA) as mobile phase B. The gradient conditions for sample elution were as follows: 2% of mobile phase B was maintained for 0.5 min; mobile phase B was linearly increased from 2% at 0.5 min to 50% at 9 min and then to 98% at 12 min, and then it was held at 98% until 13 min; finally, solvent B was decreased to 2% at 14 min. Flow rate was kept at 0.4 mL min−1, and the column temperature was fixed at 40°C. The autosampler temperature was kept at 4°C, and the volume for each injection was 2 μL. Mass data acquisition was performed in both positive and negative ESI modes by using the following operation process: ion spray voltage of 5.5 kV in the positive ESI mode and −4.0 kV in the negative ESI mode; declustering potential of 80 V in the positive ESI mode and −80 V in the negative ESI mode; source temperature of 600 °C; curtain gas of 35 psi: gas 1 (nebuliser gas) of 60 psi and gas 2 (heater gas) of 65 psi. Mass spectrometry was determined by full scan method with the range of 50-1000 m/z.
Multivariate Statistical Analysis
Mass spectra data (wiff files) were converted to mzXML files by MSConvert in Proteowizard software (version 3.0.8789). The converted raw data were further processed by XCMS software running under R (version 3.2.3) and outputted as a retention time and m/z dataset. The intensities of mass peaks for each sample were sum-normalised and Pareto-scaled using the SIMCA-P software package (version 13.0). Principal component analysis (PCA) and partial least squares-discriminant analysis (PLS-DA) with data from the 12 samples were performed to observe differences in metabolic compositions of the two potato materials. A total of 100 random permutation tests were performed to prevent overfitting of the PLS-DA model. The PLS-DA model was evaluated by R2 and Q2. R2X and R2Y indicated the fraction of the variance of x and y variable modeled in the components, whereas Q2Y denoted the predictive performance.
Screening and Identification of Biomarkers
The [p (corr)] values of all the metabolites and the first component were calculated through an S-plot of the PLS-DA model. The metabolites identified as potential markers satisfied the following criteria: (i) high confidence [|p(corr)| > 0.8] in discriminations between SD92 and SD140; (ii) mean intensities in SD92 significantly different from those in SD140 (p < 0.05); (iii) high impact intensity and explanatory ability [variable importance on projection (VIP) > 1]. The p value was calculated by an independent two-sample t-test.
The metabolites were confirmed on the basis of their exact molecular weights, and the possible empirical formulae of the metabolites were speculated (molecular weight error ≤ 20 ppm). The exact molecular weights were then used to identify potential biomarkers by mapping them in the Human Metabolome Database (http://www.hmdb.ca/), Metlin (http://metlin.scripps.edu/website), massbank (http://www.massbank.jp/) and Lipid Maps (http://www.lipidmaps.org). The metabolites were confirmed by comparing their mass spectra and retention indices to those found in the databases at a cut-off value of 85%.
Metabolite Analysis
The raw data of differential metabolites, namely the intensities of mass peak, were sum-normalised and scaled, and then a hierarchical cluster analysis (HCA) was done and a pheatmap of differential metabolites was produced in R (version 3.2.3), respectively. The intensities of mass peak of differential metabolites were sum-normalised, and then the metabolite correlation analysis was performed using cor ( ) and cor.test ( ) in R (version 3.2.3).
Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis of Differential Metabolites
In KEGG analysis, the compound numbers of differential metabolites were obtained and the differential metabolites were mapped to the corresponding KEGG pathways on the internet (https://www.genome.jp/kegg/pathway.html).
Correlation Analysis of Metabolome and Transcriptome
Pearson correlation coefficients were calculated for the differential metabolites in metabolome data of this experiment and the differentially expressed genes in transcriptome data of previous study (Liu et al. 2018). A false-positive check was performed, and the correlation was considered to be significant at a false discovery rate of p ≤ 0.05 in R (version 3.2.3).
Real-Time qRT-PCR Validation of Differentially Expressed Genes
To validate the differential genes in the network composed of these differential genes and differential metabolites, real-time qRT-PCR was conducted. Gene-specific primers are listed in Table S1. Two-microliter cDNA were used as templates in a 20-μL reaction volume using 2×SYBR Green Pro Taq HS Premix (Accurate Biotechnology, China) following the manufacturer’s instruction. The qPCR reaction was performed using 7500 Real-Time PCR (ABI, USA). The qPCR conditions were 40 cycles of 5 s at 95 °C and 30 s at 60 °C, followed by 65 to 95°C melting curve detection. Three biological replicates were analysed and 18S RNA was used as a reference gene.
Yeast One–Hybrid Assay
A yeast one–hybrid (Y1H) assay was performed using Matchmaker™ Gold Yeast One-Hybrid Library Screening System (Takara, Japan). Two sequences (196 bp and 307 bp) in the promoter of F3'5'H were cloned into the pAbAi vector. A cDNA primary library of SD92 and SD140 was constructed by use by CloneMiner II kit (Invitrogen, USA), and the primary library was inserted into the pGADT7 vector to construct a secondary library. The secondary library was transformed into the Y1H Gold strain. DNA-protein interaction was assessed based on the growth of the co-transformants on synthetic dextrose plates lacking leucine, but containing 300 ng·mL-1 aureobasidin A. Positive clones were selected by PCR, and the amplified products were detected by agarose gel electrophoresis and sequenced.
Results
Global Metabolic Differences Between Purple Potato and Its Red Mutant
Datasets obtained via UPLC–Triple-TOF–MS were further processed by PCA to compare the metabolite compositions of purple tuber and its red mutant. The results of PCA displayed that the first two principal components (PCs) explained 39.3% of the total variance in the positive ESI mode (Fig. S1a); the first two PCs explained 43.4% of the total variance in the negative ESI mode (Fig. S1b). These results indicated that SD92 and SD140 were clearly separated in the PC1 × PC2 score plots.
Furthermore, the score plots of PLS-DA model were applied to investigate the differences between SD92 and SD140 (Fig. 1a and b). The values of R2Y and Q2 in both ESI modes (Table 1) indicated the goodness of the data fit and the predictive ability of the model. The results of the PLS-DA permutation test demonstrated that all permuted Q2 values were lower than the original point of the R2 value (Fig. S2a and S2b). These results indicated that the PLS-DA models in both ESI modes were valid and reliable.
Screening and Identification of Biomarkers
In the positive ESI mode, a total of 259 mass ions were identified as the candidate biomarkers, of which 105 ions upregulated in SD92 and 154 downregulated in SD140 (Fig. 2a and b), respectively. From the negative ESI mode, there were a total 491 mass ions filtered, of which 152 and 339 ions were upregulated in SD92 and down-regulated in SD140, respectively.
In order to further identify the chemical structures of these candidate biomarkers, the MS/MS analysis of their molecular ions was also conducted with both ESI modes. The secondary mass spectra were obtained. Compared with those of the above-mentioned databases, the chemical structures of these biomarkers were identified, 10 metabolites were detected in positive ESI mode (Table 2) and 11 in negative ESI mode (Table 3), respectively.
Metabolite Correlation Analysis
To research probable discrepancies in the metabolite profiles between SD92 and SD140, HCA was performed in 12 samples. The results displayed that SD92 was clustered into one class and SD140 was clustered into another class (Fig. 3), indicating that the differential metabolites detected could effectively distinguish the two groups of samples. To further investigate the regulation mode of the anthocyanin biosynthesis branch, the associations among 19 identified metabolites were evaluated through HCA. The results showed that these metabolites could be clustered into two groups. One group contained 11 metabolites, and the relative contents of them in SD140 were higher than those in SD92. This group mainly contained flavonoids, such as pelargonidin, kaempferol-3-O-rutinoside, apigenin, and luteolin (Table 2). The other group consisted of L-glutathione, 13-HpOTrE, 4-aminobenzoic acid, laricitrin 3-glucoside, and a set of nucleotides, whose contents were lower in SD140 than in SD92.
To investigate the associations among the differential metabolites, Pearson correlation coefficient analysis was carried out. L-Glutathione, uridine monophosphate, guanosine 5'-monophosphate, and guanosine 3',5'-cyclic monophosphate were defined as Group I, and there were significantly positive correlations among them. Group II consisted of chlorogenic acid, eriodictin-7-O-glucoside, apigenin, kaempferol-7-O-glucoside, luteolin, pelargonidin, kaempferol-3-O-rutinoside, pelargonidin-3-O-glucoside, and xanthosine, and significantly positive correlations were presented among them. All nucleotides in Group I were significantly and negatively correlated with those in Group II. Pelargonidin showed a significantly positive correlation with phlorizin and nicotinamide adenine dinucleotide. Pelargonidin and pelargonidin-3-O-glucoside exhibited significantly negative correlations with 13-HpOTrE, 4-aminobenzoic acid, and laricitrin 3-glucoside. Kaempferol-3-O-rutinoside or kaempferol-7-O-glucoside was significantly and positively correlated with phlorizin, nicotinamide adenine dinucleotide, and tyrosine. Kaempferol-3-O-rutinoside was significantly and negatively correlated with L-glutathione. Laricitrin 3-glucoside was significantly and negatively correlated with phlorizin and nicotinamide adenine dinucleotide.
Metabolic Pathways Involved in Differential Metabolites
The pathways of these differential metabolites and their related synthetic precursors were analysed via KEGG analysis. The results revealed that the upregulated metabolites in SD140 were mainly related to the pathways of flavonoid biosynthesis, catecholamine biosynthesis, oxidative phosphorylation, and amino acid biosynthesis (Table 4). Flavonoid biosynthesis included flavone and flavonol biosynthesis, anthocyanin biosynthesis, apigenin biosynthesis, luteolin biosynthesis, and chlorogenic acid biosynthesis. Amino acids biosynthesis primarily comprised biosynthesis of phenylalanine, tyrosine, and tryptophan. The downregulated differential metabolites in SD140 were chiefly involved in the pathways of pyrimidine metabolism, purine metabolism, glutathione metabolism, and biosynthesis of alkaloids (Table 5).
Correlation Analysis of Metabolome and Transcriptome
In order to explore the origin of differential metabolites, a correlation analysis of metabolic pathway between the differential metabolites and the differential genes was performed. The results showed that F3'5'H and seven differential metabolites, namely, pelargonidin, pelargonidin-3-O-glucoside, apigenin, chlorogenic acid, luteolin, kaempferol-7-O-glucoside, and laricitrin were in the flavonoid biosynthesis pathway. In this metabolic pathway, the contents of pelargonidin, pelargonidin-3-O-glucoside, luteolin, chlorogenic acid, apigenin, kaempferol-7-O-glucoside increased, and laricitrin levels decreased. To investigate the genes and metabolites regulating F3'5'H expression, differential genes highly correlated with F3'5'H (r2 > 0.8) were screened. The correlations between these screened differential genes and differential metabolites were analysed and then a network was constructed.
The correlation analysis identified 63 differentially expressed genes highly correlated with F3'5'H (Table S2). These genes mainly encoded enzymes, transcription factors, and unknown proteins. The enzymes primarily included synthetase, transferase, oxidase, and hydrolase (phosphatase). The transcription factors were composed of leucine-rich repeat protein, WD repeat-containing protein, and MADS-box transcription factor.
The network revealed that 11 genes and 10 metabolites presented 76 significant correlations, including 17 positive correlations and 58 negative correlations (Table S3, Fig. 4). These genes mainly encoded serine/threonine protein kinase, fatty acid amide hydrolase, mitochondrial import receptor, UMP-CMP kinase, MADS-box transcription factor, and WD repeat-containing protein. The function of PGSC0003DMG400008074 was unknown (Table S2). The expressions of PGSC0003DMG400022867 encoding MADS-box transcription factor and BGI_novel_G000694 encoding WD repeat-containing protein were downregulated in SD140. The expression of gene encoding MADS-box transcription enhancer factor was negatively correlated with pelargonidin, luteolin, kaempferol-3-O-rutinoside, and eriodictyol-7-O-glucoside and positively correlated with uridine monophosphate. The expression of gene encoding WD repeat-containing protein was positively correlated with uridine monophosphate.
Correlation analysis of differential metabolites and differential genes. Significant correlations have R2 ≥ 0.8 and false discovery rate (FDR) < 0.05. Metabolites are represented by rectangles, and genes are denoted by circles. Correlations are indicated by connected lines. Positive correlations are blue, whereas negative correlations are red
Real-Time qRT-PCR Validation of the Genes Expression Difference
To validate the differential expressions of the genes in the network, real-time qRT-PCR of 10 genes among them was performed. The qRT-PCR result indicated that all 10 genes expressions in SD92 and SD140 were differential. And the expression pattern was accordant with the data from transcriptome (Fig. 5).
Screening of Proteins Interacting with F3'5'H Promoter
The second library plasmids were co-transformed into pAbAi-F3'5'H-I and pAbAi-F3'5'H-II yeast competent cells. After screening co-transformants, 61 and 30 positive colonies were confirmed, respectively (Fig. 6). The sequencing analysis showed that the cDNA sequences of positive colonies encoded ethylene-responsive transcription factor ERF011, gibberellin-regulated protein 9-like, and serine threonine-protein kinase, transcription factor LHW-like Bhlh, and so on.
Discussion
In this work, there are 19 differential metabolites between purple SD92 and red mutant SD140, mainly including flavonoids and nucleotides. Due to the change of colour in SD92 and SD140, we focused more on colour-related differential metabolites. Among these differential metabolites, pelargondinin is a kind of anthocyanin (Martin and Gerats 1993). In SD140, the content of pelargondinin increased. According to the anthocyanin metabolic pathway, pelargondinin can make plant tissues appear red (Holton and Cornish 1995). So the change of colour in SD92 and SD140 resulted from increased pelargonidin content. In SD140, the content of pelargondinin increased, resulting in an inrease of pelargonidin-3-O-glucoside content. In addition, in the metabolic pathway of flavonols and flavonoids, dihydrokaempferol is the upstream substrate of pelargonidin, and both kaempferol-7-O-glucoside and apigenin are closely related to dihydrokaempferol (map00941), so the increased contents of kaempferol-7-O-glucoside and apigenin led to the accumulation of pelargonidin in SD140. What is more, pelargondinin can be biosynthesised only when F3'5'H function is lost (Liu et al. 2018), and the expression of F3'5'H in SD140 significantly decreased during transcriptome analysis, so the increase of pelargondinin content in SD140 was related to the downregulated expression of F3'5'H. Laricitrin is a downstream product of F3'5'H, and the decrease in laricitrin content was related to the decrease in the expression of F3'5'H.
Pelargonidin is the key differential metabolite between SD92 and SD140. The pelargonidin content was highly correlated with the expressions of genes encoding CBL-interacting serine/threonine-protein kinase 11, UMP-CMP kinase 3-like, mitochondrial import receptor subunit TOM40-1, mitochondrial-like isoform X1, MADS-box transcription factor 2A, and MADS-box of the transcription enhancer factor 2C.
CBL-interacting serine/threonine-protein kinase is a protein kinase that catalyses protein phosphorylation. Protein kinases have a correlation with anthocyanin biosynthesis. Mitogen-activated protein (MAP) kinase kinase 6, MAP kinase kinase 9, and MAP KINASE4 are involved in regulating anthocyanin biosynthesis by MYB 75 (Wersch et al. 2018; Li et al. 2016; Luo et al. 2017). Plant sucrose-nonfermenting 1 (SNF1)-related protein kinase 1 and SNF1-related kinase 2 improve anthocyanin accumulation by MdbHLH3 and ABA, respectively (Shen et al. 2014, 2017; Liu et al. 2017). A plasma membrane-localised receptor-like protein kinase, namely, botrytis-induced kinase 1, plays a critical role in anthocyanin accumulation (Zhang et al. 2016). In this study, the gene encoding CBL-interacting serine/threonine-protein kinase had a significantly positive correlation with F3'5'H and a negative correlation with pelargonidin, and the results of yeast single-hybrid experiments showed that serine/threonine-protein kinase interacted with the promoter of F3'5'H, suggesting that CBL-interacting serine/threonine-protein kinase regulated pelargonidin biosynthesis by decreasing F3'5'H expression.
Transcription factor plays an important role in regulating anthocyanin biosynthesis (Jung et al. 2009; Damelia et al. 2014). Among the differential genes highly correlated with F3'5'H, two genes encoding MADS-box transcription enhancer factor and WD repeat-containing protein were down-regulated in SD140. As a SQUAMOSA-class MADS box transcription factor, VmTDR4 influences anthocyanin accumulation by upregulating R2R3MYB family (Jaakola et al. 2010). R2R3MYB regulates anthocyanin biosynthesis through F3′5′H (Wessinger and Rausher 2015). Therefore, MADS may regulate anthocyanin biosynthesis through F3′5′H. The present study showed that F3′5′H was positively correlated with MADS-box; hence, the decrease of F3′5′H expression in SD140 was caused by the decrease in MADS-box content. In addition, F3'5'H dominates petunidin biosynthesis (Rommens et al. 2008; Brueggemann et al. 2010); thus, the petunidin content in SD140 decreased. When neither F3'5'H nor F3'H is expressed, pelargonidin is directly biosynthesised (Holton and Cornish 1995; Liu et al. 2018). As expected, in the present study, the expression of gene encoding MADS was negatively correlated with pelargonidin content. As a WD repeat-containing protein, WD40 is in plant cytoplasm. Several WD40 repeat proteins, such as TT1 of A. thaliana, HvWD40 of barley, and StAN11 of potato, have been identified; these proteins regulate the expressions of structural genes in anthocyanin biosynthesis pathway at the transcriptional level (Brueggemann et al. 2010; Ben-Simhon et al. 2011; Strygina et al. 2017; Li et al. 2014). In the present work, F3′5′H was positively correlated with WD repeat-containing proteins, illustrating that a decrease in the content of WD repeat-containing proteins resulted in a decrease in F3′5′H expression. Then, the decrease in F3′5′H expression further led to a decrease of petunidin content and an increase of pelargonidin content in SD140. So WD40 repeat protein regulated anthocynin biosynthesis branch through F3′5′H. The results of yeast single-hybrid experiments showed that LHW bHLH interacted with the promoter of F3'5'H. bHLH can regulate the anthocyanin biosynthesis (Damelia et al. 2014; Liu et al. 2016), and has a close relationship with WD40 (Fogelman et al. 2015), so both bHLH and WD40 regulated anthocyanine biosynthesis in SD92. Metabolome analysis revealed that the expression of gene encoding WD repeat-containing proteins was positively correlated with uridine monophosphate content. The function of uridine monophosphate in regulating F3′5′H expression is worth investigating.
Conclusion
In summary, the present study identified 19 key differential metabolites, including 10 flavonoids and 4 nucleotides, which were involved in the regulation of pelargondinin biosynthesis. Ten differential metabolites among them and 11 differential genes construct a network regulating the anthocyanin biosynthesis branch. The differential genes encoding CBL-interacting serine/threonine-protein kinase, MADS-box transcription enhancer factor, and WD repeat-containing proteins negatively regulated the anthocyanin biosynthesis branch from petunidin to pelargonidin by positively regulating F3'5'H expression. The results provide the valuable insights into the regulatory mechanism of the anthocyanin biosynthesis branch in potatoes.
References
Beattie J, Crozier A, Duthie GG (2005) Potential health benefits of berries. Curr Nutr Food Sci 1(1):71–86
Ben-Simhon Z, Judeinstein S, Nadler-Hassar T, Trainin T, Bar-Ya'akov I, Borochov-Neori H, Holland D (2011) A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 234(5):865–881
Bogs J, Ebadi A, McDavid D, Robinson SP (2006) Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiol 140:279–291
Brown C, Culley D, Bonierbale M, Amoros W (2007) Anthocyanin, carotenoid content, and antioxidant values in native south American potato cultivars. Hortscience 42(7):1733–1736
Brueggemann J, Weisshaar B, Sagasser M (2010) A WD40-repeat gene from Malus × domestica is a functional homologue of Arabidopsis thaliana TRANSPARENT TESTA GLABRA1. Plant Cell Rep 29:285–294
Castellarin SD, Di Gaspero G, Marconi R, Nonis A, Peterlunger E, Paillard S, Adam-Blondon AF, Testolin R (2006) Colour variation in red grapevines (Vitis vinifera L.): genomic organisation, expression of flavonoid 3'-hydroxylase, flavonoid 3',5'-hydroxylase genes and related metabolite profiling of red cyanidin-/blue delphinidin-based anthocyanins in berry skin. BMC Genomics 7(12). https://doi.org/10.1186/1471-2164-7-12
Cavallini E, Zenoni S, Finezzo L, Guzzo F, Zamboni A, Avesani L, Tornielli GB (2014) Functional diversification of grapevine MYB5a and MYB5b in the control of flavonoid biosynthesis in a petunia anthocyanin regulatory mutant. Plant Cell Physiol 55(3):517–534
Chen S, Zhou H, Zhang G, Meng J, Deng K, Zhou W, Wang H, Wang Z, Hu N, Suo Y (2019) Anthocyanins from Lycium ruthenicum Murr. ameliorated D-galactose-induced memory impairment, oxidative stress, and neuroinflammation in adult rats. J Agric Food Chem 67(11):3140–3149
Cutanda-Perez MC, Ageorges A, Gomez C, Vialet S, Terrier N, Romieu C, Torregrosa L (2009) Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol Biol 69:633–648
Damelia V, Aversano R, Batelli G, Caruso I, Castellano Moreno M, Castro-Sanz AB, Chiaiese P, Fasano C, Palomba F, Carputo D (2014) High AN1 variability and interaction with basic helix-loop-helix co-factors related to anthocyanin biosynthesis in potato leaves. Plant J 80(3):527–540
Degu A, Hochberg U, Sikron N, Venturini L, Buson G, Ghan R, Plaschkes I, Batushansky A, Chalifa-Caspi V, Mattivi F, Delledonne M, Pezzotti M, Rachmilevitch S, Cramer GR, Fait A (2014) Metabolite and transcript profiling of berry skin during fruit development elucidates differential regulation between Cabernet Sauvignon and Shiraz cultivars at branching points in the polyphenol pathway. BMC Plant Biol 14:188. https://doi.org/10.1186/s12870-014-0188-4
Falginella L, Castellarin SD, Testolin R, Gambetta GA, Morgante M, Gaspero GD (2010) Expansion and subfunctionalisation of flavonoid 3',5'-hydroxylases in the grapevine lineage. BMC Genomics 11(1):562–562
Fogelman E, Tanami S, Ginzberg I (2015) Anthocyanin synthesis in native and wound periderms of potato. Physiol Plant 153:616–626
Galvano F, La Fauci L, Lazzarino G, Fogliano V, Ritieni A, Ciappellano S, Battistini NC, Tavazzi B, Galvano G (2004) Cyanidins: metabolism and biological properties. J Nutr Biochem 15(1):2–11
Giusti MM, Polit MF, Ayvaz H, Tay D, Manrique I (2014) Characterization and quantitation of anthocyanins and other phenolics in native Andean potatoes. J Agric Food Chem 62(19):4408–4416
Han KH, Sekikawa M, Shimada K, Hashimoto M, Naoto H, Noda T, Tanaka H, Fukushima M (2006) Anthocyanin-rich purple potato flake extract has antioxidant capacity and improves antioxidant potential in rats. Br J Nutr 96(6):1125–1134
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071–1083
Hu C, Li Q, Shen X, Quan S, Lin H, Duan L, Wang Y, Luo Q, Qu G, Han Q, Lu Y, Zhang D, Yuan Z, Shi J (2016) Characterization of factors underlying the metabolic shifts in developing kernels of colored maize. Sci Rep 6:35479
Jaakola L, Poole M, Jones MO, Kamarainen-KarppinenT KJJ, Hohtola A, Haggman H, Fraser PD, Manning K, King GJ, Thomson H, Seymour GB (2010) A SQUAMOSA MADS Box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol 153(4):1619–1629
Jeong ST, Goto-Yamamoto N, Hashizume K, Esaka M (2006) Expression of the flavonoid 3'-hydroxylase and flavonoid 3',5'-hydroxylase genes and flavonoid composition in grape (Vitis vinifera). Plant Sci 170:61–69
Jung CS, GriYths HM, De Jong DM, Cheng SP, Bodis M, Kim TS, De Jong WS (2009) The potato developer (D) locus encodes an R2R3 MYB transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin. Theor Appl Genet 120(1):45–57
Li W, Wang B, Wang M (2014) Cloning and characterization of a potato StAN11 gene involved in anthocyanin biosynthesis regulation. J Integr Plant Biol 56(4):364–372
Li S, Wang W, Gao J, Yin K, Wang R, Wang C, Petersen M, Mundy J, Qiu J (2016) MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 28(11):2866–2883
Liu Y, Wang K, Espley RV, Wang L, Yang H, Yu B (2016) Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J Exp Bot 67:2159–2176
Liu X, An X, Liu X, Hu D, Wang X, You C, Hao Y (2017) MdSnRK1.1 interacts with MdJAZ18 to regulate sucrose-induced anthocyanin and proanthocyanidin accumulation in apple. J Exp Bot 68(11):2977–2990
Liu F, Yang Y, Gao J, Ma C, Bi Y (2018) A comparative transcriptome analysis of a wild purple potato and its red mutant provides insight into the mechanism of anthocyanin transformation. PLoS One 13(1):e0191406
Long H, Zhang F, Wang H, Yang W, Hou H, Yu J, Liu B (2018) Mulberry anthocyanins improves thyroid cancer progression mainly by inducing apoptosis and autophagy cell death. Kaohsiung J Med Sci 34(5):255–262
Lu Q, Yang Q (2006) cDNA cloning and expression of anthocyanin biosynthetic genes in wild potato (Solanum pinnatisectum). Afr J Biotechnol 5:811–818
Luo J, Wang X, Feng L, Li Y, He J (2017) The mitogen-activated protein kinase kinase 9 (MKK9) modulates nitrogen acquisition and anthocyanin accumulation under nitrogen-limiting condition in Arabidopsis. Biochem Biophys Res Commun 487(3):539–544
Martin C, Gerats T (1993) Control of pigment biosynthesis genes during petal development. Plant Cell 5:1253–1264
Mei Y, Xie H, Liu S, Zhu J, Zhao S, Wei C (2021) Metabolites and Transcriptional Profiling Analysis Reveal the Molecular Mechanisms of the Anthocyanin Metabolism in the "Zijuan" Tea Plant (Camellia sinensis var. assamica). J Agric Food Chem 69(1):414–427
Momose M, Nakayama M, Itoh Y, Umemoto N, Toguri T, Ozeki Y (2013) An active hAT transposable element causing bud mutation of carnation by insertion into the flavonoid 3'-hydroxylase gene. Mol Gen Genomics 288:175–184
Olsen KM, Hehn A, Jugde H, Slimestad R, Larbat R, Bourgaud F (2010) Identification and characterisation of CYP75A31, a new flavonoid 3′5′-hydroxylase, isolated from Solanum lycopersicum. BMC Plant Biol 10:10–21
Prior RL (2004) Absorption and metabolism of anthocyanins: potential health effects. In: Meskin MS, Bidlack WR, Davies AJ, Lewis DS, Randolph RK (eds) Phytochemicals-mechanism of action, 1st edn. CRC Press, Boca Raton, FL, pp 1–19
Rommens CM, Richael CM, Yan H, Navarre DA, Ye J, Krucker M, Swords K (2008) Engineered native pathways for high kaempferol and caffeoylquinate production in potato. Plant Biotechnol J 6:870–886
Saito T, Nishida M, Saito M, Tanabe A, Eitsuka T, Yuan S, Ikekawa N, Nishida H (2016) The fruit of Acanthopanax senticosus (Rupr. et Maxim.) Harms improves insulin resistance and hepatic lipid accumulation by modulation of liver adenosine monophosphate-activated protein kinase activity and lipogenic gene expression in high-fat diet-fed obese mice. Nutr Res 36(10):1090–1097
Shen X, Zhao K, Liu L, Zhang K, Yuan H, Liao X, Wang Q, Guo X, Li F, Li T (2014) A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng (Prunus avium L.). Plant Cell Physiol 55:862–880
Shen X, Guo X, Zhao D, Zhang Q, Jiang Y, Wang Y, Peng X, Wei Y, Zhai Z, Zhao W, Li T (2017) Cloning and expression profiling of the PacSnRK2 and PacPP2C gene families during fruit development, ABA treatment, and dehydration stress in sweet cherry. Plant Physiol Biochem 119:275–285
Strygina KV, Börner A, Khlestkina EK (2017) Identification and characterization of regulatory network components for anthocyanin synthesis in barley aleurone. BMC Plant Biol 17(suppl1):184
Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, Noji M, Yamazaki M, Saito KO (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42(2):218–235
Wang Z, Cui Y, Vainstein A, Chen S, Ma H (2017a) Regulation of fig (Ficus carica L.) fruit color: metabolomic and transcriptomic analyses of the flavonoid biosynthetic pathway. Front Plant Sci 8:1990
Wang L, Sun X, Weiszmann J, Weckwerth W (2017b) System-level and granger network analysis of integrated proteomic and metabolomic dynamics identifies key points of grape berry development at the interface of primary and secondary metabolism. Front Plant Sci 8:1066
Wersch RV, Gao F, Zhang Y (2018) Mitogen-activated protein kinase kinase 6 negatively regulates anthocyanin induction in Arabidopsis. Plant Signal Behav 13(10):e1526000
Wessinger CA, Rausher MD (2015) Ecological transition predictably associated with gene degeneration. Mol Biol Evol 32(2):347–354
Whang SS, Um WS, Song IJ, Lim PO, Choi K, Park KW, Kang KW, Choi MS, Koo JC (2011) Molecular Analysis of Anthocyanin Biosynthetic Genes and Control of Flower Coloration by Flavonoid 3′,5′-Hydroxylase (F3′5′H) in Dendrobium moniliforme. J Plant Biol 54(3):209–218
Yan F, Zhang J, Zhang L, Zheng X (2016) Mulberry anthocyanin extract regulates glucose metabolism by promotion of glycogen synthesis and reduction of gluconeogenesis in human HepG2 cells. Food Funct 7(1):425–433
Zhang H, Huang L, Hong Y, Song F (2016) BOTRYTIS-INDUCED KINASE1, a plasma membrane-localised receptor-like protein kinase, is a negative regulator of phosphate homeostasis in Arabidopsis thaliana. BMC Plant Biol 16(1):152
Acknowledgements
This work was supported by National Natural Science Foundation of China (31901589) and Modern Agriculture Industrial Technology System of Shandong Province (SDAIT-16-05) sponsored by Shandong Provincial Department of Agriculture.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information

Fig. S1
Score plots of the PCA model obtained from purple potato and its red mutant. (a) Positive ESI mode. (b) Negative ESI mode. (PNG 76 kb)

Fig. S1
(PNG 195 kb)

Fig. S2
Permutation test of the PLS-DA model. (a) Positive ESI mode. (b) Negative ESI mode. (PNG 75 kb)

Fig. S2
(PNG 75 kb)
Table S1
List of primers used for validation of the differently expressed genes (DOCX 12 kb)
Table S2
Differentially expressed genes screened with significant correlation with F3'5'H expression (DOCX 19 kb)
Table S3
Interaction values between differential metabolites and differential genes. (DOCX 16 kb)
Table S4
Sequence analysis of protein interacting with the promoter of F3'5'H (DOCX 22 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, F., Chen, G., Zhang, Y. et al. A Comparative Analysis of Metabolome Reveals the Regulation of the Anthocyanin Biosynthesis Branch in Potatoes. Potato Res. 66, 831–850 (2023). https://doi.org/10.1007/s11540-022-09586-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11540-022-09586-5