Abstract
Introduction
The burden of stroke in patients with hypertension is very high, and its prediction is critical.
Objectives
We aimed to use plasma lipidomics profiling to identify lipid biomarkers for predicting incident stroke in patients with hypertension.
Methods
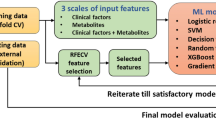
This was a nested case-control study. Baseline plasma samples were collected from 30 hypertensive patients with newly developed stroke, 30 matched patients with hypertension, 30 matched patients at high risk of stroke, and 30 matched healthy controls. Lipidomics analysis was performed by ultrahigh-performance liquid chromatography–tandem mass spectrometry, and differential lipid metabolites were screened using multivariate and univariate statistical methods. Machine learning methods (least absolute shrinkage and selection operator, random forest) were used to identify candidate biomarkers for predicting stroke in patients with hypertension.
Results
Co-expression network analysis revealed that the key molecular alterations of the lipid network in stroke implicate glycerophospholipid metabolism and choline metabolism. Six lipid metabolites were identified as candidate biomarkers by multivariate statistical and machine learning methods, namely phosphatidyl choline(40:3p)(rep), cholesteryl ester(20:5), monoglyceride(29:5), triglyceride(18:0p/18:1/18:1), triglyceride(18:1/18:2/21:0) and coenzyme(q9). The combination of these six lipid biomarkers exhibited good diagnostic and predictive ability, as it could indicate a risk of stroke at an early stage in patients with hypertension (area under the curve = 0.870; 95% confidence interval: 0.783–0.957).
Conclusions
We determined lipidomic signatures associated with future stroke development and identified new lipid biomarkers for predicting stroke in patients with hypertension. The biomarkers have translational potential and thus may serve as blood-based biomarkers for predicting hypertensive stroke.





Similar content being viewed by others
Data Availability
The raw lipidomic data generated in this study have been deposited in the Open Archive for Miscellaneous Data (OMIX) database (https://www.cncb.ac.cn/) with accession Number OMIX004817. Other data that support the findings of this study are available from the corresponding author upon request.
Abbreviations
- AUC:
-
Area under the curve
- BMI:
-
Body mass index
- CV:
-
Coefficient of variation
- CHD:
-
Coronary heart disease
- ChE:
-
Cholesteryl ester
- CHSCs:
-
Community health service centers
- Co:
-
Coenzyme
- DG:
-
Diglyceride
- FC:
-
Fold change
- HDL-C:
-
Hig-density lipoprotein cholesterol
- LASSO:
-
Least absolute shrinkage and selection operator
- LDL-C:
-
Low-density lipoprotein cholesterol
- LOESS:
-
Locally estimated scatterplot smoothing
- LPC:
-
Lysophosphatidylcholine
- MG:
-
Monoglycerde
- PC:
-
Phosphatidylcholine
- PE:
-
Phosphatidylethanolamine
- PG:
-
Phosphatidylglycerol
- PhSM:
-
Phytosphingosine
- PIP:
-
Phosphatidylinositol
- PLS-DA:
-
Partial least squares discriminant analysis
- PS:
-
Phosphatidylserine
- QC:
-
Quality control
- ROC:
-
Receiver operating characteristic
- RF:
-
Random forest
- SiE:
-
Sitosteryl ester
- SM:
-
Sphingomyelin
- STROBE:
-
Strengthening the Reporting of Observational studies in Epidemiology
- TC:
-
Total cholesterol
- TG:
-
Triglyceride
- UPLC-MS/MS:
-
Ultra-high performance liquid chromatography-tandem mass spectrometry
- VIP:
-
Variable importance in projection
- WGCNA:
-
Weighted gene co-expression network analysis
References
Amarenco, P., Goldstein, L. B., Szarek, M., Sillesen, H., Rudolph, A. E., Callahan, A., et al. (2007). Effects of intense low-density lipoprotein cholesterol reduction in patients with Stroke or transient ischemic Attack: The Stroke Prevention by Aggressive reduction in cholesterol levels (SPARCL) trial. Stroke, 38(12), 3198–3204. https://doi.org/10.1161/STROKEAHA.107.493106. 3rd.
Au, A. (2018). Metabolomics and Lipidomics of ischemic Stroke. Advances in Clinical Chemistry, 85, 31–69. https://doi.org/10.1016/bs.acc.2018.02.002
Bergholt, M. S., Serio, A., McKenzie, J. S., Boyd, A., Soares, R. F., Tillner, J., et al. (2018). Correlated Heterospectral Lipidomics for Biomolecular Profiling of Remyelination in multiple sclerosis. ACS Central Science, 4(1), 39–51. https://doi.org/10.1021/acscentsci.7b00367
Bose, G., Healy, B. C., Lokhande, H. A., Sotiropoulos, M. G., Polgar-Turcsanyi, M., Anderson, M., et al. (2022). Early predictors of Clinical and MRI outcomes using least Absolute Shrinkage and Selection Operator (LASSO) in multiple sclerosis. Annals of Neurology, 92(1), 87–96. https://doi.org/10.1002/ana.26370
Buonacera, A., Stancanelli, B., & Malatino, L. (2019). Stroke and Hypertension: An Appraisal from Pathophysiology to Clinical Practice. Current Vascular Pharmacology, 17(1), 72–84. https://doi.org/10.2174/1570161115666171116151051
Cipolla, M. J., Liebeskind, D. S., & Chan, S. L. (2018). The importance of comorbidities in ischemic Stroke: Impact of Hypertension on the cerebral circulation. Journal of Cerebral Blood flow and Metabolism: Official Journal of the International Society of Cerebral Blood Flow and Metabolism, 38(12), 2129–2149. https://doi.org/10.1177/0271678X18800589
del Zoppo, G. J., Poeck, K., Pessin, M. S., Wolpert, S. M., Furlan, A. J., Ferbert, A., et al. (1992). Recombinant tissue plasminogen activator in acute thrombotic and embolic Stroke. Annals of Neurology, 32(1), 78–86. https://doi.org/10.1002/ana.410320113
Dong, M. X., Hu, L., Wei, Y. D., & Chen, G. H. (2021). Metabolomics profiling reveals altered lipid metabolism and identifies a panel of lipid metabolites as biomarkers for Parkinson’s Disease related anxiety disorder. Neuroscience Letters, 745, 135626. https://doi.org/10.1016/j.neulet.2021.135626
Drescher, C., Buchwald, F., Ullberg, T., Pihlsgård, M., Norrving, B., & Petersson, J. (2022). Epidemiology of First and recurrent ischemic Stroke in Sweden 2010–2019: A Riksstroke Study. Neuroepidemiology, 56(6), 433–442. https://doi.org/10.1159/000527373
Eichelmann, F., Sellem, L., Wittenbecher, C., Jäger, S., Kuxhaus, O., Prada, M., et al. (2022). Deep lipidomics in human plasma: Cardiometabolic Disease Risk and Effect of Dietary Fat Modulation. Circulation, 146(1), 21–35. https://doi.org/10.1161/CIRCULATIONAHA.121.056805
Fahy, E., Sud, M., Cotter, D., & Subramaniam, S. (2007). LIPID MAPS online tools for lipid research. Nucleic Acids Research, 35, W606–612. https://doi.org/10.1093/nar/gkm324. Web Server issue.
GBD 2019 Stroke Collaborators Global. (2021). Global, regional, and national burden of Stroke and its risk factors, 1990–2019: A systematic analysis for the global burden of Disease Study 2019. The Lancet Neurology, 20(10), 795–820. https://doi.org/10.1016/S1474-4422(21)00252-0
Giles, C., Takechi, R., Lam, V., Dhaliwal, S. S., & Mamo, J. (2018). Contemporary lipidomic analytics: Opportunities and pitfalls. Progress in Lipid Research, 71, 86–100. https://doi.org/10.1016/j.plipres.2018.06.003
Graessler, J., Schwudke, D., Schwarz, P. E., Herzog, R., Shevchenko, A., & Bornstein, S. R. (2009). Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PloS One, 4(7), e6261. https://doi.org/10.1371/journal.pone.0006261
Guo, X., Li, Z., Zhou, Y., Yu, S., Yang, H., Zheng, L., et al. (2019). Metabolic Profile for Prediction of Ischemic Stroke in Chinese Hypertensive Population. Journal of Stroke and Cerebrovascular Diseases: The Official Journal of National Stroke Association, 28(4), 1062–1069. https://doi.org/10.1016/j.jstrokecerebrovasdis.2018.12.035
Han, X. (2016). Lipidomics for studying metabolism. Nature Reviews Endocrinology, 12(11), 668–679. https://doi.org/10.1038/nrendo.2016.98
Han, L., Wu, Q., Wang, C., Hao, Y., Zhao, J., Zhang, L., et al. (2015). Homocysteine, ischemic Stroke, and Coronary Heart Disease in Hypertensive patients: A Population-Based, prospective cohort study. Stroke, 46(7), 1777–1786. https://doi.org/10.1161/STROKEAHA.115.009111
Heo, J. H., Nam, H. S., Kim, Y. D., Choi, J. K., Kim, B. M., Kim, D. J., & Kwon, I. (2020). Pathophysiologic and therapeutic perspectives based on Thrombus histology in Stroke. Journal of Stroke, 22(1), 64–75. https://doi.org/10.5853/jos.2019.03440
Jové, M., Mauri-Capdevila, G., Suárez, I., Cambray, S., Sanahuja, J., Quílez, A., et al. (2015). Metabolomics predicts Stroke recurrence after transient ischemic Attack. Neurology, 84(1), 36–45. https://doi.org/10.1212/WNL.0000000000001093
Ke, C., Zhu, X., Zhang, Y., & Shen, Y. (2018). Metabolomic characterization of Hypertension and dyslipidemia. Metabolomics: Official Journal of the Metabolomic Society, 14(9), 117. https://doi.org/10.1007/s11306-018-1408-y
Kessler, R. M. (2003). Imaging methods for evaluating brain function in man. Neurobiology of Aging, 24 Suppl(1), 21–35. https://doi.org/10.1016/s0197-4580(03)00047-2. discussion S37-39.
Khoury, S., Beauvais, A., Colas, J., Saint-Martin Willer, A., Perros, F., Humbert, M., et al. (2022). Lipidomic Profile Analysis of Lung Tissues revealed Lipointoxication in Pulmonary Veno-Occlusive Disease. Biomolecules, 12(12), 1878. https://doi.org/10.3390/biom12121878
Krug, S., Kastenmüller, G., Stückler, F., Rist, M. J., Skurk, T., Sailer, M., et al. (2012). The dynamic range of the human metabolome revealed by challenges. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 26(6), 2607–2619. https://doi.org/10.1096/fj.11-198093
Kulkarni, H., Meikle, P. J., Mamtani, M., Weir, J. M., Barlow, C. K., Jowett, J. B., et al. (2013). Plasma lipidomic profile signature of Hypertension in Mexican American families: Specific role of diacylglycerols. Hypertension, 62(3), 621–626. https://doi.org/10.1161/HYPERTENSIONAHA.113.01396
Kulkarni, H., Mamtani, M., Blangero, J., & Curran, J. E. (2017). Lipidomics in the study of Hypertension in metabolic syndrome. Current Hypertension Reports, 19(1), 7. https://doi.org/10.1007/s11906-017-0705-6
Langfelder, P., & Horvath, S. (2008). WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559. https://doi.org/10.1186/1471-2105-9-559
Liu, P., Li, R., Antonov, A. A., Wang, L., Li, W., Hua, Y., et al. (2017). Discovery of Metabolite biomarkers for Acute ischemic Stroke progression. Journal of Proteome Research, 16(2), 773–779. https://doi.org/10.1021/acs.jproteome.6b00779
Liu, J., Tang, L., Lu, Q., Yu, Y., Xu, Q. G., Zhang, S., et al. (2022). Plasma quantitative lipid profiles: Identification of CarnitineC18:1-OH, CarnitineC18:2-OH and FFA (20:1) as novel biomarkers for pre-warning and prognosis in Acute Myocardial Infarction. Frontiers in Cardiovascular Medicine, 9, 848840. https://doi.org/10.3389/fcvm.2022.848840
Ma, Y., Li, L., Niu, Z., Song, J., Lin, Y., Zhang, H., & Du, G. (2016). Effect of recombinant plasminogen activator timing on thrombolysis in a novel rat embolic Stroke model. Pharmacological Research, 107, 291–299. https://doi.org/10.1016/j.phrs.2016.03.030
Meikle, P. J., & Christopher, M. J. (2011). Lipidomics is providing new insight into the metabolic syndrome and its sequelae. Current Opinion in Lipidology, 22(3), 210–215. https://doi.org/10.1097/MOL.0b013e3283453dbe
Niu, Z., Wu, Q., Luo, Y., Wang, D., Zheng, H., Wu, Y., et al. (2022). Plasma lipidomic subclasses and risk of Hypertension in Middle-aged and Elderly Chinese. Phenomics (Cham Switzerland), 2(5), 283–294. https://doi.org/10.1007/s43657-022-00057-y
O’Donnell, M. J., Xavier, D., Liu, L., Zhang, H., Chin, S. L., Rao-Melacini, P., et al. (2010). Risk factors for ischaemic and intracerebral haemorrhagic Stroke in 22 countries (the INTERSTROKE study): A case-control study. Lancet, 376(9735), 112–123. https://doi.org/10.1016/S0140-6736(10)60834-3
Pei, G., Chen, L., & Zhang, W. (2017). WGCNA Application to Proteomic and Metabolomic Data Analysis. Methods in Enzymology, 585, 135–158. https://doi.org/10.1016/bs.mie.2016.09.016
Pullagurla, S. R., Witek, M. A., Jackson, J. M., Lindell, M. A., Hupert, M. L., Nesterova, I. V., et al. (2014). Parallel affinity-based isolation of leukocyte subsets using microfluidics: Application for Stroke diagnosis. Analytical Chemistry, 86(8), 4058–4065. https://doi.org/10.1021/ac5007766
Sun, D., Tiedt, S., Yu, B., Jian, X., Gottesman, R. F., Mosley, T. H., et al. (2019). A prospective study of serum metabolites and risk of ischemic Stroke. Neurology, 92(16), e1890–e1898. https://doi.org/10.1212/WNL.0000000000007279
Tomita, Y., Sakata, S., Arima, H., Yamato, I., Ibaraki, A., Ohtsubo, T., et al. (2021). Relationship between casual serum triglyceride levels and the development of Hypertension in Japanese. Journal of Hypertension, 39(4), 677–682. https://doi.org/10.1097/HJH.0000000000002693
von Elm, E., Altman, D. G., Egger, M., Pocock, S. J., Gøtzsche, P. C., & Vandenbroucke, J. P. (2007). The strengthening the reporting of Observational studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. PLoS Medicine, 4(10), e296. https://doi.org/10.1371/journal.pmed.0040296
Wang, C. Y., Chen, Z. W., Zhang, T., Liu, J., Chen, S. H., Liu, S. Y., et al. (2014). Elevated plasma homocysteine level is associated with ischemic Stroke in Chinese hypertensive patients. European Journal of Internal Medicine, 25(6), 538–544. https://doi.org/10.1016/j.ejim.2014.04.011
Wang, Z., Chen, Z., Zhang, L., Wang, X., Hao, G., Zhang, Z., et al. (2018). Status of Hypertension in China: Results from the China Hypertension Survey, 2012–2015. Circulation, 137(22), 2344–2356. https://doi.org/10.1161/CIRCULATIONAHA.117.032380
Wen, B., Mei, Z., Zeng, C., & Liu, S. (2017). metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinformatics, 18(1), 183. https://doi.org/10.1186/s12859-017-1579-y
Yoon, J. H., Seo, Y., Jo, Y. S., Lee, S., Cho, E., Cazenave-Gassiot, A., et al. (2022). Brain lipidomics: From functional landscape to clinical significance. Science Advances, 8(37), eadc9317. https://doi.org/10.1126/sciadv.adc9317
Yu, Y., Wen, X., Lin, J. G., Liu, J., Liang, H. F., Lin, S. W., et al. (2023). Identification of three potential novel biomarkers for early diagnosis of acute ischemic Stroke via plasma lipidomics. Metabolomics: Official Journal of the Metabolomic Society, 19(4), 32. https://doi.org/10.1007/s11306-023-01990-3
Zhang, J., Su, X., Qi, A., Liu, L., Zhang, L., Zhong, Y., et al. (2021). Metabolomic profiling of fatty acid biomarkers for intracerebral Hemorrhage Stroke. TALANTA, 222, 121679. https://doi.org/10.1016/j.talanta.2020.121679
Zhao, T., Yan, Q., Wang, C., Zeng, J., Zhang, R., Wang, H., et al. (2023). Identification of serum biomarkers of ischemic Stroke in a Hypertensive Population based on Metabolomics and Lipidomics. Neuroscience, 533, 22–35. https://doi.org/10.1016/j.neuroscience.2023.09.017
Zhong, C., Lu, Z., Che, B., Qian, S., Zheng, X., Wang, A., et al. (2021). Choline Pathway nutrients and metabolites and Cognitive Impairment after Acute ischemic Stroke. Stroke, 52(3), 887–895. https://doi.org/10.1161/STROKEAHA.120.031903
Zhou, M., Wang, H., Zeng, X., Yin, P., Zhu, J., Chen, W., et al. (2019). Mortality, morbidity, and risk factors in China and its provinces, 1990–2017: A systematic analysis for the global burden of Disease Study 2017. Lancet, 394(10204), 1145–1158. https://doi.org/10.1016/S0140-6736(19)30427-1
Zhou, W., Li, S., Sun, G., Song, L., Feng, W., Li, R., et al. (2022). Early warning of ischemic Stroke based on Atherosclerosis Index Combined with serum markers. The Journal of Clinical Endocrinology and Metabolism, 107(7), 1956–1964. https://doi.org/10.1210/clinem/dgac176
Acknowledgements
We thank all authors for their contributions to the article.
Funding
This study was supported by the grants from the National Natural Science Foundation of China (82173648); the Natural Science Foundation of Guangdong Province (2022A1515011273); the Shenzhen science and technology project (JCYJ20210324125810024); the Shenzhen Nanshan District Science and Technology Bureau (2020075); the NingboKey Support Medical Discipline (Grant No.2022-F22); the Public Welfare Foundation of Ningbo (2021S108); Medical Scientific Research Foundation of Zhejiang Province; China (2022RC253 and 2021RC028); Zhejiang Provincial Public Service and Application Research Foundation; China (LGC22H260005); Key Program of Ningbo Natural Science Foundation; China (2022J271); Ningbo Leading Top Talent Training Project (2022RC-LJ-01); Internal Fund of Ningbo Institute of Life and Health Industry; University of Chinese Academy of Sciences (2020YJY0212); Ningbo Health Technology Project (2022Y30); Ningbo Natural Science Foundation(2022J275); Project of NINGBO Leading Medical& Health Discipline (2022-B12); the Medical and Health Science and Technology Project in Zhejiang province (2023KY1136 and 2024KY1589); the Ningbo Health Branding Subject Fund (PPXK2018-01) and the HwaMei Reasearch Foundation of Ningbo No.2 Hospital (2021HMKY14, 2022HMKY12 and 2023HMZD01).
Author information
Authors and Affiliations
Contributions
Conceptualization, CYW and LYH; Methodology, JJZ, RJZ and HMR; Formal Analysis, RJZ and LYH; Investigation, JJZ, SX, HW and YNJ; Resources, HMR and CYW; Data Curation, JJZ and TZ; Writing – Original Draft Preparation, JJZ and TZ; Writing – Review & Editing, JJZ, LYH and CYW. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All procedures followed in this study were in accordance with the ethical standards of the Ethics Committee of the Shenzhen Nanshan Centre for Chronic Disease Control (LL20190003) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zeng, J., Zhang, R., Zhao, T. et al. Plasma lipidomic profiling reveals six candidate biomarkers for the prediction of incident stroke in patients with hypertension. Metabolomics 20, 13 (2024). https://doi.org/10.1007/s11306-023-02081-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-023-02081-z