Abstract
Large-artery atherosclerosis (LAA) is a leading cause of cerebrovascular disease. However, LAA diagnosis is costly and needs professional identification. Many metabolites have been identified as biomarkers of specific traits. However, there are inconsistent findings regarding suitable biomarkers for the prediction of LAA. In this study, we propose a new method integrates multiple machine learning algorithms and feature selection method to handle multidimensional data. Among the six machine learning models, logistic regression (LR) model exhibited the best prediction performance. The value of area under the receiver operating characteristic curve (AUC) was 0.92 when 62 features were incorporated in the external validation set for the LR model. In this model, LAA could be well predicted by clinical risk factors including body mass index, smoking, and medications for controlling diabetes, hypertension, and hyperlipidemia as well as metabolites involved in aminoacyl-tRNA biosynthesis and lipid metabolism. In addition, we found that 27 features were present among the five adopted models that could provide good results. If these 27 features were used in the LR model, an AUC value of 0.93 could be achieved. Our study has demonstrated the effectiveness of combining machine learning algorithms with recursive feature elimination and cross-validation methods for biomarker identification. Moreover, we have shown that using shared features can yield more reliable correlations than either model, which can be valuable for future identification of LAA.
Similar content being viewed by others
Introduction
Large-artery atherosclerosis (LAA) is a pathological condition characterized by the formation of chronic plaques in arteries, which can lead to obstructed blood flow and resulting in ischemic injury. LAA is a multifactorial disease responsible for 20–30% of ischemic stroke cases1. Clinically, non-invasive examinations such as ultrasound, computed tomography (CT), and magnetic resonance angiography (MRA) are typically used to confirm the diagnosis. However, these tests are often expensive and time-consuming, and their accuracy may be dependent on the skill level of the technician performing the exam. Therefore, there is an urgent clinical need to identify novel and more efficient biomarkers for predicting the risk of LAA, which can be achieved through the general blood tests.
Well-known risk factors include age, gender, and family history of stroke, hypertension, diabetes, hyperlipidemia, obesity, alcohol consumption, and tobacco smoking2. Studies have also found that endothelial dysfunction and the resulting inflammatory response may lead to compromised endothelial integrity and plaque formation3,4,5,6,7,8. Altered metabolism is a hallmark of acute myocardial ischemia, providing more real-time cell signaling information than other clinical symptoms9,10,11. Several pathways, particularly cholesterol, purine, pyrimidine, and ceramide pathways12,13,14, are found to be altered when atherosclerosis occurs. These molecules were considered being the novel biomarkers and therapeutic targets for LAA.
In addition to the complexity and heterogeneity of LAA pathology, the dynamic nature of metabolism also makes traditional statistical methods ineffective for such large and complex data sets. Machine learning (ML) approaches have shown promising in diagnosis improvement, risk prediction, and disease treatment for chronic cardiovascular diseases based on lifestyle15, biochemical testing16, electrocardiograms17, medical imaging18, and genetic, genomic, and proteomic biomarkers19. For example, a study in India used ML algorithms to automatically identify and quantify carotid artery plaques in MRI scans. They achieved 91.41% accuracy in LAA classification using Random Forest (RF)20. Another research used routine clinical data to develop a model to predict the risk of carotid plaque progression in patients with asymptomatic carotid stenosis. They found that logistic regression (LR) could provide the best predictive ability of AUC at 0.80921. In metabolomics biomarker discovery, seven lipoprotein-focused metabolites have been identified by the top features of lasso LR and random forest machine learning models. Leda et al. found that for metabolic profiles, logistic regression achieved a maximum accuracy of 0.822. Song et al. even developed a novel multi-metabolite predictive model to predict response to statin therapy in patients with atherosclerosis. They identified RA-specific abnormalities in remitted patients after PCI dominated by alternations in lipid biochemical pathways, including sphingolipid, phospholipid, eicosanoid, and fatty acid oxidation. The AUC and accuracy were up to 0.89 and 0.90, respectively23.
Although these studies have demonstrated a good performance in biomarker discovery and disease prediction, their limited AUC and accuracy hinder their scalability for clinical use. To further improve the performance, we propose a new method integrates multiple ML algorithms and feature selection method to handle multidimensional data. Different strengths and weaknesses of the algorithms are considered to find the best fit model, and the feature selection method identifies informative and relevant features to improve model generalization and reduce overfitting. Notably, we taken into consideration the importance of shared features for disease across different models. These features with strong predictive power for disease can be selected as candidate biomarkers for further research.
We found (1) The combination of clinical factor and metabolite profile provides stability to data set shifts; (2) with feature selection method we improved the model performance from an AUC of 0.89 to 0.92; (3) the shared features had predictive power equivalent to 67 features, suggesting their clinical importance in identifying patients with LAA. In this study, we attempted to develop a new biomarker discovery method which may help identify LAA less costly and more efficient.
Methods
Participants and study design
From 2010 to 2015, consecutive ischemic stroke patients with extracranial LAA were recruited according to the following inclusion criteria: (1) cerebral angiography including digital subtraction, magnetic resonance or computed tomographic angiogram exhibiting evidence of the extracranial common and internal carotid artery having ≥ 50% diameter stenosis according to NASCET criteria24; (2) stable neurological condition during blood sample collection; (3) no acute illness, such as infection or inflammation, at the time of blood sample collection; and (4) a modified Rankin Scale score of less than 3. Normal controls were recruited from the neurology outpatient department. Normal controls were defined as those with (1) no history of stroke and coronary artery disease, (2) brain magnetic resonance or computed tomographic angiogram exhibiting < 50% diameter stenosis at bilateral intracranial and extracranial carotid arteries, and (3) no acute illness during blood sample collection.
The exclusion criteria were (1) exhibiting systemic diseases, such as hypothyroidism or hyperthyroidism, decompensated liver cirrhosis, acute kidney injury, or systemic lupus erythematosus and (2) having cancer and other serious illnesses during recruitment. This study was approved by the Institutional Review Board of Linkou Chang Gung Memorial Hospital (revised approval numbers: 201506352B0C501 and 202000552B0C601). All the participants signed informed consent forms before being recruited into this study.
Venous blood samples and clinical profiles were collected at recruitment of normal controls and LAA patients in stationary condition. Blood for metabolomics analysis was stored in sodium citrate tubes and centrifuged (10 min, 3000 rpm at 4 °C) within an hour after collection. Plasma was aliquoted into separate polypropylene tubes and stored at − 80 °C freezer. The measurement of metabolites was done following our previous method (Lin CN et al., 2021) using the targeted Absolute IDQ®p180 kit (Biocrates Life Science, AG, Innsbruck, Austria) which can quantify 194 endogenous metabolites from 5 classes of compound. The assay was performed by using a Waters Acquity Xevo TQ-S instrument (Waters, Milford, MA, USA). The level of metabolite was obtained by using the Biocrates®® MetIDQ™ software.
Data preprocessing and parameters of machine learning models
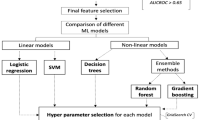
The workflow of this study is shown in Fig. 1. The data preprocessing steps involved missing data handling, label encoding, and participant grouping. Open-source specialized packages for Python, including Pandas24,25, NumPy26, scikit-learn27, Matplotlib28, Seaborn29, TableOne30, and SciPy31, were applied. We used the mean imputation method of the Useful package32 in R to obtain the missing values for each variable. After converting the categorical variables into dummy variables, we used 80% of the data set for model training/validation (tenfold cross-validation training set, n = 287) and the remaining 20% for performance testing (external validation set, n = 72). Three scales of input features including clinical factors, metabolites and clinical factors + metabolites were adopted using six machine learning models: logistic regression (LR), support vector machine (SVM), decision tree, random forest (RF), extreme gradient boosting (XGBoost), and gradient boosting (Supplementary Methods).
The flowchart of ML models used for the prediction of LAA. AUC area under the receiver operating characteristic curve; CV cross-validation, LAA large-artery atherosclerosis; ML machine learning; RFECV recursive feature elimination with cross-validation; SVM support vector machine; XGBoost extreme gradient boosting.
LR is a supervised learning technique used to address classification issues and determine the likelihood of a binary (yes/no) occurrence. Whatever the variable is dichotomous or categorical, the logistic function use a S-shaped curve to transform data into a value between 0 and 1 for classification issues33.
SVM was first proposed by Corinna et al.34. Processing nonlinear, small-sample, and high-dimensional pattern recognition problems with SVM provides various benefits. It offers a great generalization ability for unknown samples because the partitioning hyperplane may ensure that the extreme solution is a global optimal solution rather than a local minimal value and has a solid theoretical foundation35.
Decision tree algorithm is a common type of machine learning algorithm in which decisions are made according to a tree structure. A decision tree typically has a root node, a number of internal nodes, and a number of leaf nodes. The root node includes all of the samples, and each node's samples are separated into subnodes based on the outcomes of an attribute test. The sequence of decision tests corresponds to the route from the root node to the last leaf node35,36.
RF is an extension of the bagging method37, which is a typical ensemble learning method. Bagging often entails processing chores using a straightforward voting system. A decision tree algorithm serves as the foundation learner for RF, and during decision tree training, random attribute selection is included. For a variety of real-world data, RF offers reliable performance, and is easily understood35. It has shown good performance in applications like disease prediction, gene selection, and picture recognition38,39,40.
XGBoost is a novel gradient boosting ensemble learning method. In this method, machine learning is implemented under the gradient boosting framework with high efficiency, flexibility, and portability41. Tree boosting is an efficient and widely used machine learning method that is a type of boosted ensemble learning42. The second-order Taylor expansion of the loss function is used by the XGBoost model, and a regularization function is added to this expansion to strike a compromise between the model's complexity and loss function reduction. This approach attempts to avoid overfitting to some extent by looking for the overall ideal solution35. The gradient tree boosting algorithm used by XGBoost is to increase its speed and accuracy.
Gradient boosting is a fast and accurate machine-learning-based prediction method that is particularly well suited for large and complicated datasets. Gradient boosting redefines boosting as a numerical optimization problem with the objective of minimizing the loss function by incorporating a weak learner via gradient descent. In order to reduce the overall error of the strong learner, the contribution of each weak learner to the final prediction is based on a gradient optimization procedure. Gradient boosting focuses on existing underperforming learners43.
In the models of this study, we set the maximum number of iterations as 3,000 and added a penalty term (L2) to the loss function in our LR model. For our SVM model, the radial basis function was used as the kernel function, the regularization parameter (C) was 1.0, and the class weight was set as “balanced.” For the decision tree model, the maximum depth of a tree was 6, it was determined through optimization procedures and based on previous studies. For the RF and gradient boosting algorithms, default parameter settings were used. For the XGBoost algorithm, we used the tree construction algorithm (tree_method) as “hist”, and nodes with the highest loss change were added to the tree (grow_policy).
Feature selection through recursive feature elimination with cross-validation
For reduction of the number of input variables of the machine learning models, we used the recursive feature elimination (RFE) with cross-validation (RFECV) method to identify the important features in this study (Fig. 2). The RFECV method has received considerable research attention because of its robustness44. RFE is a greedy algorithm based on the packing model. The RFECV algorithm starts from a complete feature set, and its performance metric is the prediction accuracy or area under the receiver operating characteristic curve (AUC) of the classifier. At the end of an iteration, the least relevant features are eliminated. The most relevant features are then sorted and extracted. RFE involves the extraction of feature subsets according to the feature ranking table generated on the basis of the aforementioned evaluation metrics35.
Model evaluation metrics
Baseline metabolite levels and clinical factors were presented in terms of mean ± standard deviation. Categorical variables were expressed as absolute and percentage frequencies. The Python 3.7 software package and scikit-learn toolkit were adopted, and the default settings were applied to train with the LR, SVM, decision tree, RF, XGBoost, and gradient boost algorithms.
We used the four metrics including accuracy, AUC, recall, and precision to evaluate the performance of the machine learning models.
where TP denotes true positives, FP represents false positives, TN denotes true negatives, and FN refers to false negatives.
The input data was split into a training set and an external validation set, following an 8:2 ratio. Subsequently, the training set was divided into k equal parts for k-fold internal cross-validation. During each of the k iterations, one part of the training set was designated for internal validation, while the remaining k − 1 parts were employed for training the models. This approach facilitated thorough evaluation and model training using the training sets. Lastly, the performance of the models was evaluated using the external validation set. This procedure was repeated until each of the k subsets had been served as the validation set. The average of the k performance measurements was the cross-validated performance45. In this study, we conducted internal stratified tenfold cross-validation in the training set to estimate the performance of the models46. And the final performance of the models was evaluated using the external validation set. The RFECV algorithm was used to determine the contributions of features to the predictions classified into the LAA and “control” categories.
Mean absolute difference (MAD)
The mean absolute difference (MAD) is a statistical measure used to quantify the average discrepancy between individual data points and a reference value47. It is calculated by taking the absolute difference between each data point and the reference value, then computing the average of these absolute differences. MAD provides valuable insights into the dispersion or variability of a dataset, allowing researchers to assess the magnitude of differences from a central reference point.
Ethics statement
All methods described in this study were carried out in accordance with relevant guidelines and regulations. The studies involving human participants were reviewed and approved by the Institutional Review Board of Linkou Chang Gung Memorial Hospital (revised approval numbers: 201506352B0C501 and 202000552B0C601) and informed consent was obtained from all participants prior to their inclusion in the study. The study also adhered to the principles outlined in the Declaration of Helsinki and the International Conference on Harmonization-Good Clinical Practice (ICH-GCP) guidelines.
Results
Patient population and demographics
There were 359 people who participated in the study, with 176 of them were LAA, and the remaining 183 were normal control (Table 1 and Supplementary Fig. S1). The mean age of the LAA cohort was 64 years (range: 58–69 years), while the control cohort was 61 years (range: 56–66 years). The two groups of individuals who participated in the study did not differ in sex, family history of stroke, chronic kidney disease, and most anthropometric measures. However, there were significant differences between the two groups in hypertension, diabetes mellitus, and usage of long-term medication (p < 0.001). Compared with the normal controls, the patients with LAA were almost twice as likely as normal controls to exhibit unhealthy lifestyle behaviors such as smoking (40.4% vs. 73.3%, p < 0.001) and alcohol usage (24.4% vs. 42.0%, p < 0.001). Moreover, the patients with LAA had higher levels of homocysteine and creatinine but lower levels of high-density/low-density lipoprotein and total cholesterol (all p < 0.01) than the normal controls.
In the analysis of serum metabolites, significant differences were observed between the two groups of 68 out of the 164 analyzed metabolites (41.5%; Table 2). Patients with LAA exhibited lower levels of 34 phosphatidylcholines (PCs, 50.0% in 68) than the normal controls, whereas higher levels of amino acids (6 out of 21; 28.6%), biogenic amines (two out of six; 33.3%), lysoPCs (1 out of 12; 8.3%), and other markers (three out of seven; 42.9%) were observed in the LAA group (all; p < 0.05). Additionally, 17 acylcarnitines showed differences between the two groups, with 13 acylcarnitines exhibiting lower levels and 4 showing higher levels in the LAA group (p < 0.05). Of the 11 sphingomyelins examined, only SMOHC141 showed a significant difference between the two groups (p = 0.006). The examined metabolites are presented in Supplementary Table 1 and Fig. S2.
Performance of the adopted models in predicting LAA using three scales of input features
The performance of the 6 predictive models is presented in Table 3, using the 10-fold cross validation. After training, all of these models exhibited high AUC values but low precision, recall, and accuracy. When the training was performed on clinical factors, metabolites and clinical factors + metabolites, the mean performances in clinical factors were accuracy: 0.76 ± 0.18, AUC: 0.84 ± 0.15, recall: 0.75 ± 0.28, precision: 0.75 ± 0.20; those in metabolites were accuracy: 0.70 ± 0.13, AUC: 0.76 ± 0.12, recall: 0.68 ± 0.20, precision: 0.69 ± 0.16; those in clinical factors + metabolites were accuracy: 0.77± 0.13, AUC: 0.83 ± 0.12, recall: 0.74 ± 0.23, precision: 0.77 ± 0.16. We found that the models trained with clinical factors had similar performance to models trained with clinical factors + metabolites in predicting LAA, but the best fit algorithm was different for each input feature scale. When considering only clinical factors for training the models, the RF model exhibited the best LAA prediction performance in all the 4 metrics (AUC: 0.90 ± 0.14, accuracy: 0.82 ± 0.17, recall: 0.81 ± 0.26, and precision: 0.82 ± 0.21). When considering only metabolites factors, the LR model exhibited the best AUC value of 0.81 ± 0.10 with accuracy: 0.73 ± 0.12, recall: 0.72 ± 0.12, and precision: 0.72 ± 0.17. When combining both clinical factors and metabolites factors, the LR model exhibited the best AUC value of 0.89 ± 0.12 with accuracy: 0.78 ± 0.15, recall: 0.77 ± 0.26, and precision: 0.77 ± 0.17.
To test the robustness, we evaluated the model performance by the AUCs within the external validation set. The mean AUC for the 6 models tested by clinical factors, metabolites and clinical factors + metabolites were 0.84 ± 0.04, 0.81 ± 0.07, and 0.86 ± 0.08, respectively (Fig. 3). In models tested by clinical factors, the SVM exhibited the best AUC value of 0.88 [Fig. 3A]. However, in models tested by metabolites, all AUC values decreased, especially the decision tree model [AUC: 0.65, Fig. 3B]. The XGBoost exhibited the best AUC value of 0.86 [Fig. 3B]. In models tested by clinical factors + metabolites, the AUC of the decision tree model increased to 0.67, and the others also increased to over 0.89 [Fig. 3C]. Gradient boost model exhibited the best AUC value of 0.92 [Fig. 3C]. The mean absolute difference (MAD) between training and testing dataset were then calculated (Table 4). The MAD of logistic regression model was 0.02, which was the lowest among the 6 models. Indicating, LR model consistently exhibited the best or second-best performance for different input scales.
Receiver operating characteristic curves for the 6 machine learning models evaluated with the external validation set using 3 scales of input features: (A) clinical factors, (B) metabolites, and (C) combination of clinical factors and metabolites. SVM support vector machine, XGBoost extreme gradient boosting.
Feature selection by using the RFECV method
Tables 3 and 4 show using both clinical factors and metabolites as features provided the best AUC and robustness over the 6 models. Figure 4 illustrates the selection process to obtain the best subset of features from clinical factors + metabolites (193 features) in each model (Supplementary Table 2). The results indicated that five models achieved an AUC value over 0.87 when using the RFECV method (Fig. 4). Only the decision tree model exhibited an AUC value of 0.71. Among the 6 models, the LR model used the least number of features but achieved the best performance (AUC = 0.90).
RFECV curves for the 6 adopted ML models. The red dot-line represents the number of features required to attain the highest AUC value. AUC area under the receiver operating characteristic curve; ML machine learning; RFECV recursive feature elimination with cross-validation; SVM support vector machine; XGBoost extreme gradient boosting.
Since the AUC score did not increase substantially when more than 62 features were selected, 62 features were selected to train a new LR model. These features comprised 15 clinical factors and 47 metabolites (Supplementary Table 3). A tenfold cross-validation was performed on the training set to estimate the generalization capability of the LR model. Similar AUC values were obtained for each part of the training set, indicating that no overfitting occurred [Fig. 5A]. The mean AUC value of the LR model was 0.96 ± 0.03 which was better than the other models trained with different original inputs and methods. When the external validation set was used, an AUC value of 0.92 was obtained for the LR model [Fig. 5B] with accuracy = 0.82 [Fig. 5C], with six patients being misclassified as belonging to the normal cohort and six members of the normal cohort being misclassified as patients with LAA.
Feature selection using the RFECV method for the LR algorithm (62 features): (A) receiver operating characteristic curves for tenfold cross-validation on the training set, (B) receiver operating characteristic curves on the external validation set, and (C) confusion matrix for the external validation set. FN false negative; FP false positive; LR logistic regression; NPV negative predictive value; RFECV recursive feature elimination with cross-validation; TN true negative; TP true positive.
Shared features could improve the performance of the 6 models
Since different models have different advantages in data classification which affects the feature selection results of RFECV method, we attempted to use a Venn diagram to find shared features identified from 5 models through RFECV (Fig. 6) to understand how feature sharing could affect the adopted models. The decision tree model was excluded because of its poor predictive power in predicting LAA patients in both training and testing dataset. We found that 27 features were shared among the 5 models (LR, SVM, RF, XGBoost, and the gradient boosting). Of these features, 11 were clinical factors and 16 were serum metabolites (Supplementary Table 4).
After training, LR model still exhibited the best AUC (0.93 ± 0.10), followed by SVM model (AUC: 0.91 ± 0.07), RF model (AUC: 0.90 ± 0.13), XGBoost model (AUC: 0.90 ± 0.10) and gradient boost model (AUC: 0.90 ± 0.11) (Table 5). For the external validation set, except for decision tree (AUC: 0.55), the other 5 models exhibited good performance with AUC ≈ 0.9 [Fig. 7A]. The LR model could correctly classified 58 out of 72 patients (accuracy: 0.81), with 3 LAA patients being misclassified as normal controls and 11 normal controls being misclassified as LAA patients [Fig. 7B].
Performance of the 6 predictive models when using the 27 shared features for training: (A) receiver operating characteristic curves of the five models for the external validation set and (B) confusion matrix for the external validation set when using the LR model. FN false negative; FP false positive; NPV negative predictive value; SVM support vector machine; TN true negative; TP true positive; XGBoost extreme gradient boosting.
Performance comparison with other research
Several classification algorithms have been developed in various studies for predicting cardiovascular risk using different input data types and methods. Table 6 provides a comparison of their accuracy and AUC values. However, significant progress had not been made until 2020 when Du et al. applied six machine learning methods to electronic health records (EHR) data for predicting cardiovascular risk, achieving an AUC of 0.94, accuracy of 0.87, and Recall of 0.82. In 2022, Huang et al. enhanced a logistic regression model with multivariate methods and clinical data, achieving an AUC of 0.93 for identifying carotid atherosclerosis (CAS). Nevertheless, clinical factors or EHR data typically reflect specific physiological markers and may not fully represent the overall health status of the patient, which could limit their ability to predict certain complex diseases. Additionally, there may be overlapping blood test values among different diseases, which could increase the risk of confusion and misinterpretation when predicting complex conditions. Therefore, in this study, we utilized clinical data combined with metabolomics data to develop a predictive model that could more accurately reflect the actual physical state of the patient.
Discussion
In this study, we considered common vascular risk factors and serum metabolites for predicting extracranial LAA. The cross-comparison results of 18 models (3 scales of input features in 6 commonly used machine learning algorithms) showed the combination of clinical factors and metabolites had better predictive performance across different training algorithms, and the robustness could be observed through using the external validation set [Fig. 3C] which indicated LR was the best fit algorithm for LAA prediction (AUC: 0.89).
Previous study in Kazakh Chinese suggested that using clinical factors with SVM (AUC: 0.86) and LR (AUC: 0.87) algorithms had better performance in predicting cardiovascular disease48. Another study in Northern China found that training the models using clinical factors alone could limit the model performance with the AUC range between 0.55 and 0.67 in LR, RF and SVM models in predicting acute ischemic stroke49. However, many studies have pointed out that tree-based algorithms such as RF and gradient boosting were much more useful than regression-based algorithms in predicting large vessel occlusion with advanced measurements and pretreatment scoring methods50,51,52. For example, Alanazi et al. had trained a RF model with an AUC of 0.97 in Caucasian53; Nishi and his colleagues had trained a RF model with an AUC of 0.85 in Japanese54 ; and Wang et al. had trained a gradient boosting model with an AUC of 0.83 in Han Chinese55. Our study demonstrated the performance metrics could be improved if a feature selection step was added and both clinical factors and metabolites could be combined for disease prediction. Also, the performance could be improved if the feature number was reduced from 193 to 62 in the LR model with the mean AUC arranged from 0.89 ± 0.12 [Table 3 and Fig. 3C] to 0.96 ± 0.03 [Fig. 5A].
The 62 features selected through RFECV comprised 15 clinical factors and 47 metabolites (Supplementary Table 3). Among these clinical factors, some may be related to atherosclerosis. For instance, age usually reflects lifestyle and physical condition56. Studies have found that older individuals exhibit higher collagen deposition and vascular stiffness50, which might contribute to subsequent hypertension and the expression of several proinflammatory molecules that ultimately promote atherosclerosis51. High levels of blood pressure, cholesterol, and glucose may aggravate the risk of atherosclerosis by increasing the rate of plaque buildup and causing endothelial injury or dysfunction52,57. The balance among serum lipids, platelets, hemodynamic factors, and the blood vessel wall may influence the development of atherosclerosis58.
As for the 47 metabolites, the association with atherosclerosis was less discussed. We then conducted a pathway analysis on the 47 metabolites by using MetaboAnalyst v5.0 (Supplementary Table 5)59. The pathway enrichment analysis indicated that these metabolites were associated with aminoacyl-tRNA biosynthesis and lipid metabolism (p < 0.05). We found that the levels of half of the examined PCs were significantly lower in the patients with LAA than normal controls. The reasons for this result remain unclear. Studies have examined the prognostic values of PCs and lysoPCs and have observed a lower level in patients with coronary and peripheral arterial disease60,61. Moxon et al. suggested that a lower level of serum PC might lead to an increased rate of low-density lipoprotein oxidation, increased foam cell formation, and reduced cholesterol efflux62. Moreover, amino acid metabolism has strong relationships with several types of cardiovascular disease63. For example, valine, leucine, and isoleucine can ameliorate cell metabolic processes through mitochondrial biogenesis, influence macrophage foam cells, and alter lipid metabolism64. Tryptophan levels are identified to be lower in patients with atheromatous plaque or coronary heart disease than healthy individuals65. Many metabolites of tryptophan and arginine play a role in regulating athero-inflammation and atherogenesis66. Evidence suggests that disturbances in the balance and transmission of immune signaling might also affect LAA67.
This study is the first to integrate multi-model testing and the RFECV method in training and testing to create LAA prediction model. We eliminated irrelevant features, improved the learning accuracy of the models, and determined the best fitting model for our dataset. We found that sharing relevant features between models provided additional class-specific information that helped reduce the computational complexity and improve the recall rate efficiently. We used the RFECV method to identify specific features for LAA prediction. Even though we had trained a good LR model to predict LAA based on 62 features. To simplify the prediction model, we removed the decision tree and selected the 27 features shared among LR, SVM, RF, XGBoost, and gradient boosting. The performance indicated that using these 27 features could improve the 4 performance metrics to > 0.80 compared to the models using 193 features. However, if focused on the LR model, using the 27 features had a higher recall but lower precision compared to using the 62 features identified through RFECV. Our study may suggest using the 27 features in LR model could be a suitable test for screening LAA67.
LAA carries a high risk of cerebrovascular disease and results in worse outcome than other ischemic stroke subtypes1. The benefit of our study is that we developed a simple algorithm to screen LAA by using clinical factors and a simple blood test for metabolite profile instead of using a complicated neuroimaging study. Our study may help to early predict and prevent the progression of LAA. There is also potential that the use of machine learning may help to set up algorithm to solve clinical issues to improve the healthcare system. The predictive models created by machine learning can be applied in other medical conditions to improve disease diagnosis and management.
However, this study had several limitations. First, this was a single-center-based retrospective study, and external validation is required to determine the optimal model’s stability and population drift. Second, statistical problems might occur when machine learning is used on small data sets68. Our data set was balanced between the patient and control groups, and the results of tenfold cross-validation on an external validation set indicated that our model provided nearly unbiased performance estimates. Nevertheless, further large-scale cohort estimation is required before the clinical application of the model. Third, the metabolites were analyzed using a commercial kit (the Absolute IDQ®p180 kit; Biocrates Life Science, AG, Innsbruck, Austria). Different quantitative methods have different resolutions; thus, the universal usage of the optimal model might not be possible.
In conclusion, our study indicated that a high-accuracy LAA prediction model can be developed by integrating multi-model testing and the RFECV method. Compared to clinical factors or metabolites alone, data on a combination of appropriate clinical factors and metabolites provide a real-time understanding of an individual’s LAA status. The machine learning process used in this study can be adopted by researchers in future studies to construct high-performance prediction models for other complex diseases.
For future application, the LR model demonstrates accurate prediction of LAA thrombosis when compared to other scoring methods. This model can effectively assess risks in patients with LAA. Additionally, our study recognizes the significance of specific metabolite biomarkers in predicting atherosclerosis. It is our hope that these findings and recommendations will greatly contribute to optimizing treatment strategies for patients with LAA and hold immense significance in the prevention of LAA.
Data availability
The datasets generated and/or analyzed during the current study are available in the Google drive repository: https://drive.google.com/file/d/1KL-8Vtz4AFk342WZq_TReGReBVInxXAv/view?usp=share_link.
References
Ko, Y., et al. MRI-based Algorithm for Acute Ischemic Stroke Subtype Classification. 2014(2287–6391 (Print)).
Cole, J. W. Large Artery Atherosclerotic Occlusive Disease. 2017(1538–6899 (Electronic)).
Young, J. L., U. Libby P Fau-Schönbeck, & U. Schönbeck. Cytokines in the pathogenesis of atherosclerosis. 2002(0340–6245 (Print)).
Chapman, M. J. From pathophysiology to targeted therapy for atherothrombosis: a role for the combination of statin and aspirin in secondary prevention. 2007(0163–7258 (Print)).
Stoll, G., & Bendszus, M. Inflammation and atherosclerosis: Novel insights into plaque formation and destabilization. 2006(1524–4628 (Electronic)).
Emsley, H. C. & Tyrrell, P. J. Inflammation and infection in clinical stroke. 2002(0271–678X (Print)).
Libby, P. Inflammation in atherosclerosis. 2012(0028–0836 (Print)).
Elkind, M. S. Inflammation, atherosclerosis, and stroke. 2006(1074–7931 (Print)).
Bodi, V., et al. Metabolomics in the diagnosis of acute myocardial ischemia. 2013(1937–5395 (Electronic)).
Mas, S., et al. Local non-esterified fatty acids correlate with inflammation in atheroma plaques of patients with type 2 diabetes. 2010(1939–327X (Electronic)).
Paik, M. J., et al. The free fatty acid metabolome in cerebral ischemia following human mesenchymal stem cell transplantation in rats. 2009(1873–3492 (Electronic)).
Yu, B., et al. Genome-wide association study of a heart failure related metabolomic profile among African Americans in the Atherosclerosis Risk in Communities (ARIC) study. 2013(1098–2272 (Electronic)).
Walsh, M. C. et al. Impact of geographical region on urinary metabolomic and plasma fatty acid profiles in subjects with the metabolic syndrome across Europe: the LIPGENE study. 2014(1475–2662 (Electronic)).
Yap, I. K., et al. Metabolome-wide association study identifies multiple biomarkers that discriminate north and south Chinese populations at differing risks of cardiovascular disease: INTERMAP study. 2010(1535–3907 (Electronic)).
Ahmad, S., et al. Assessment of Risk Factors and Biomarkers Associated With Risk of Cardiovascular Disease Among Women Consuming a Mediterranean Diet. 2018(2574–3805 (Electronic)).
Shrivastava, A. K. et al. C-reactive protein, inflammation and coronary heart disease. Egypt. Heart J. 67(2), 89–97 (2015).
Bent Re Fau-Wheeler, M.T., et al. Systematic Comparison of Digital Electrocardiograms From Healthy Athletes and Patients With Hypertrophic Cardiomyopathy. 2015(1558–3597 (Electronic)).
Arsenault, B. J., et al. Visceral adipose tissue accumulation, cardiorespiratory fitness, and features of the metabolic syndrome. 2007(0003–9926 (Print)).
Ngo, D., et al. Aptamer-Based Proteomic Profiling Reveals Novel Candidate Biomarkers and Pathways in Cardiovascular Disease. 2016(1524–4539 (Electronic)).
Latha, S. et al. Performance analysis of machine learning and deep learning architectures on early stroke detection using carotid artery ultrasound images. Front. Aging Neurosci. 13, 828214 (2021).
Fan, J. et al. The prediction of asymptomatic carotid atherosclerosis with electronic health records: a comparative study of six machine learning models. BMC Med. Inform. Decis. Mak. 21(1), 115 (2021).
Coelewij, L. et al. Serum metabolomic signatures can predict subclinical atherosclerosis in patients with systemic lupus erythematosus. Arterioscler. Thromb. Vasc. Biol. 41(4), 1446–1458 (2021).
Cui, S. et al. Machine learning identifies metabolic signatures that predict the risk of recurrent angina in remitted patients after percutaneous coronary intervention: A multicenter prospective cohort study. Adv. Sci. (Weinh) 8(10), 2003893 (2021).
Barnett, H. J., et al. Benefit of carotid endarterectomy in patients with symptomatic moderate or severe stenosis. North American Symptomatic Carotid Endarterectomy Trial Collaborators. 1998(0028–4793 (Print)).
McKinney, W. Data Structures for Statistical Computing in Python. 2010.
Harris, C. R., et al. Array programming with NumPy. 2020(1476–4687 (Electronic)).
Pedregosa, F., et al. Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Hunter, J. D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9(3), 90–95 (2007).
Waskom, M., Botvinnik, O., O’Kane, D., Hobson, P., Lukauskas, S., Gemperline, D. C., Augspurger, T., Halchenko, Y., Cole, J. B., Warmenhoven, J., de Ruiter, J., Pye, C., Hoyer, S., Vanderplas, J., Villalba, S., Kunter, G., Quintero, E., Bachant, P., Martin, M., Qalieh, A. mwaskom/seaborn: v0.8.1 (September 2017) (v0.8.1). Zenodo. (2017).
Pollard, T. A.-O., et al. tableone: An open source Python package for producing summary statistics for research papers. 2018(2574–2531 (Electronic)).
Virtanen, P., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. 2020(1548–7105 (Electronic)).
Lander, J. P., Useful: A Collection of Handy, Useful Functions. R package version 1.2.6. 2018.
Peng, C.-Y.J., Lee, K. L. & Ingersoll, G. M. An introduction to logistic regression analysis and reporting. J. Educ. Res. 96, 3–14 (2002).
Vapnik, C. A. Support vector networks. Mach. Learn. 20(3), 273–297 (1995).
Chang, W. et al. A Machine-Learning-Based Prediction Method for Hypertension Outcomes Based on Medical Data. LID—https://doi.org/10.3390/diagnostics9040178 [doi] LID—178. 2019(2075–4418 (Print)).
Badr Hssina, A. M., Ezzikouri, M. Erritali, M. A comparative study of decision tree ID3 and C4.5. Int. J. Adv. Comput. Sci. Appl. (IJACSA) (2014).
Bauer, E. & Kohavi, R. An empirical comparison of voting classification algorithms: bagging, boosting, and variants. Mach. Learn. 36(1), 105–139 (1999).
Díaz-Uriarte, R., Alvarez de Andrés, S. Gene selection and classification of microarray data using random forest. 2006(1471–2105 (Electronic)).
Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 26(1), 217–222 (2005).
Gray, K. R. et al. Random forest-based similarity measures for multi-modal classification of Alzheimer's disease. 2013(1095–9572 (Electronic)).
Chen, T. and Guestrin, C. XGBoost: A Scalable Tree Boosting System. 2016. arXiv:1603.02754.
Nielsen, D. Tree Boosting with XGBoost—Why Does XGBoost Win "Every" Machine Learning Competition? 2016.
Le, N. Q., Nguyen, T. T., & Ou, Y. Y. Identifying the molecular functions of electron transport proteins using radial basis function networks and biochemical properties. 2017(1873–4243 (Electronic)).
Ding, Y., Wilkins, D. Improving the performance of SVM-RFE to select genes in microarray data. 2006(1471–2105 (Electronic)).
Berrar, D., Cross-Validation (2018).
Govindarajan, M. & Chandrasekaran, R. Evaluation of k-nearest neighbor classifier performance for direct marketing. Expert Syst. Appl. 37(1), 253–258 (2010).
Pham-Gia, T. & Hung, T. L. The mean and median absolute deviations. Math. Comput. Model. 34(7), 921–936 (2001).
Jiang, Y., et al. Cardiovascular Disease Prediction by Machine Learning Algorithms Based on Cytokines in Kazakhs of China. 2021(1179–1349 (Print)).
Cui, J., et al. Machine Learning-Based Model for Predicting Incidence and Severity of Acute Ischemic Stroke in Anterior Circulation Large Vessel Occlusion. 2021(1664–2295 (Print)).
Wang, J. C. & Bennett, M. Aging and atherosclerosis. Circ. Res. 111(2), 245–259 (2012).
Tesauro, M., et al. Arterial ageing: from endothelial dysfunction to vascular calcification. 2017(1365–2796 (Electronic)).
Dzau, V. J. Mechanism of the interaction of hypertension and hypercholesterolemia in atherogenesis: The effects of antihypertensive agents. 1988(0002–8703 (Print)).
Alanazi, E.A.-O., Abdou, A.A.-O. & Luo, J.A.-O.X. Predicting Risk of Stroke From Lab Tests Using Machine Learning Algorithms: Development and Evaluation of Prediction Models. 2021(2561–326X (Electronic)).
Nishi, H., et al. Predicting Clinical Outcomes of Large Vessel Occlusion Before Mechanical Thrombectomy Using Machine Learning. 2019(1524–4628 (Electronic)).
Wang, J.A.-O., et al. Prediction of large vessel occlusion for ischaemic stroke by using the machine learning model random forests. 2022(2059–8696 (Electronic)).
Ohya, Y., et al. Causes of ischemic stroke in young adults versus non-young adults: A multicenter hospital-based observational study. 2022(1932–6203 (Electronic)).
Lusis, A. J. Atherosclerosis. Nature 407(6801), 233–241 (2000).
Packham, M. A. & Mustard, J. F. The role of platelets in the development and complications of atherosclerosis. 1986(0037–1963 (Print)).
Xia, J., et al. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. 2009(1362–4962 (Electronic)).
Kuksis, A., Fau-Myher, J. J., et al. Decreased plasma phosphatidylcholine/free cholesterol ratio as an indicator of risk for ischemic vascular disease. 1982(0276–5047 (Print)).
Paapstel, K., et al. Inverse relations of serum phosphatidylcholines and lysophosphatidylcholines with vascular damage and heart rate in patients with atherosclerosis. 2018(1590–3729 (Electronic)).
Moxon, J. V., et al. Baseline serum phosphatidylcholine plasmalogen concentrations are inversely associated with incident myocardial infarction in patients with mixed peripheral artery disease presentations. 2017(1879–1484 (Electronic)).
Zaric, B. L., et al. Atherosclerosis Linked to Aberrant Amino Acid Metabolism and Immunosuppressive Amino Acid Catabolizing Enzymes. 2020(1664–3224 (Electronic)).
Grajeda-Iglesias, C. & Aviram, M. Specific Amino Acids Affect Cardiovascular Diseases and Atherogenesis via Protection against Macrophage Foam Cell Formation: Review Article. LID—https://doi.org/10.5041/RMMJ.10337 [doi] LID—e0022. 2018(2076–9172 (Print)).
Baldo-Enzi, G. et al. Tryptophan and atherosclerosis. In Recent Advances in Tryptophan Research 429–432 (Springer, 1996).
Li, Y., et al. Regulating the balance between the kynurenine and serotonin pathways of tryptophan metabolism. 2017(1742–4658 (Electronic)).
Sun, L., et al. Metabolic Reprogramming in Immune Response and Tissue Inflammation. 2020(1524–4636 (Electronic)).
Kokol, P.A.-O., Kokol, M., Zagoranski, S. Machine learning on small size samples: A synthetic knowledge synthesis. 2022(2047–7163 (Electronic)).
Weng, S.A.-O., et al. Can machine-learning improve cardiovascular risk prediction using routine clinical data? 2017(1932–6203 (Electronic)).
Terrada, O., et al. Classification and Prediction of atherosclerosis diseases using machine learning algorithms. in 2019 5th International Conference on Optimization and Applications (ICOA) (2019).
Lyu, J., et al. A potent risk model for predicting new-onset acute coronary syndrome in patients with type 2 diabetes mellitus in Northwest China. 2020(1432–5233 (Electronic)).
Sakellarios, A. I., et al. Prediction of atherosclerotic disease progression combining computational modelling with machine learning. in 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) (2020).
Du, Z.A.-O., et al. Accurate Prediction of Coronary Heart Disease for Patients with Hypertension From Electronic Health Records with Big Data and Machine-Learning Methods: Model Development and Performance Evaluation. 2020(2291–9694 (Print)).
Brajesh Kumar, D. H. M. Comprehensive analysis of atherosclerosis disease prediction using machine learning. Ann. Roman. Soc. Cell Biol. 25(4), 17962–17975 (2021).
Huang, G. et al. Development and validation of a carotid atherosclerosis risk prediction model based on a Chinese population. 2022(2297–055X (Print)).
Chen, Z. et al. Prediction of atherosclerosis using machine learning based on operations research. 2022(1551–0018 (Electronic)).
Funding
The study was funded by China Medical University and China Medical University Hospital, grant numbers: DMR-112-124, DMR-111-227, MOST 110-2314-B-039-010-MY2, MOST 111-2321-B-039-005, MOST 111-2321-B-030-004 and MOST 111-2622-8-039-001-IE, and Linkou Chang Gung Memorial Hospital, grant numbers: CMRPG3E2131, CMRPG3F2211-2, CMRPG3K2351.
Author information
Authors and Affiliations
Contributions
T.-H.S. and C.-C.W. contributed equally to this work (Co-first author). K.-C.H. and T.-H.L. contributed equally to the manuscript (Co-corresponding author). Y.-L.W. assists to the research recommendation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, TH., Wang, CC., Wu, YL. et al. Machine learning approaches for biomarker discovery to predict large-artery atherosclerosis. Sci Rep 13, 15139 (2023). https://doi.org/10.1038/s41598-023-42338-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-42338-0
- Springer Nature Limited
This article is cited by
-
Unlocking potential biomarkers bridging coronary atherosclerosis and pyrimidine metabolism-associated genes through an integrated bioinformatics and machine learning approach
BMC Cardiovascular Disorders (2024)
-
Mechanism exploration and biomarker identification of glycemic deterioration in patients with diseases of the exocrine pancreas
Scientific Reports (2024)