Abstract
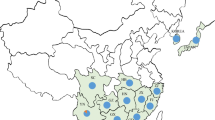
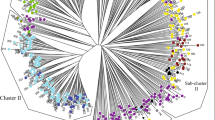
Pear (Pyrus L.) is one of the most important fruits in temperate regions. Even in the mountainous border region of Tibet, Sichuan, and Yunnan Provinces (China), a variety of highly adapted pear landraces were discovered in Tibetan Autonomous Prefecture and were named “Zangli pears.” Here, we analyzed genetic diversity of 29 genotypes of Zangli distributed in Tibetan Region, together with 30 genotypes of sand pear (P. pyrifolia) in border regions, and 8 genotypes of P. ussuriensis as out groups, using simple sequence repeats (SSR) markers. Twenty-eight SSR markers displayed high polymorphism with a total of 202 alleles. Cluster analysis revealed a close genetic relationship between Zangli and sand pears, while population structure analysis revealed that gene flow between the western and eastern side of the Jin-sha River was restricted by geographic isolation. Diversity statistics for five geographic groups implied a migration event from Sichuan and Yunnan Provinces to the border region of western Sichuan, eastern Tibet, and northwestern Yunnan Provinces. Six specific genetic alleles in Zangli pear landraces might be related to their adaption to severe environmental conditions.




Similar content being viewed by others
References
Bailey LH (1919) The standard cyclopedia of horticulture vol 2. Macmillan
Bao L, Chen K, Zhang D, Cao Y, Yamamoto T, Teng Y (2007) Genetic diversity and similarity of pear (Pyrus L.) cultivars native to East Asia revealed by SSR (simple sequence repeat) markers. GENET RESOUR CROP EV 54:959–971
Baraket G, Chatti K, Saddoud O, Abdelkarim AB, Mars M, Trifi M, Hannachi AS (2011) Comparative assessment of SSR and AFLP markers for evaluation of genetic diversity and conservation of fig, Ficus carica L., genetic resources in Tunisia. PLANT MOL BIOL REP 29:171–184
Bittencourt J, Sebbenn AM (2007) Patterns of pollen and seed dispersal in a small, fragmented population of the wind-pollinated tree Araucaria angustifolia in southern Brazil. Heredity 99:580–591
Campoy JA, Ruiz D, Egea J, Rees DJG, Celton JM, Martínez-Gómez P (2011) Inheritance of flowering time in apricot (Prunus armeniaca L.) and analysis of linked quantitative trait loci (QTLs) using simple sequence repeat (SSR) markers. PLANT MOL BIOL REP 29:404–410
Challice JS, Westwood MN (1973) Numerical taxonomic studies of the genus Pyrus using both chemical and botanical characters. Bot J Linn Soc 67:121–148
Chen H et al (2015) Construction of a high-density simple sequence repeat consensus genetic map for pear (Pyrus spp.). PLANT MOL BIOL REP 33:316–325
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Dos Santos ARF, Ramos-Cabrer AM, Díaz-Hernández MB, Pereira-Lorenzo S (2011) Genetic variability and diversification process in local pear cultivars from northwestern Spain using microsatellites. Tree Genet Genomes 7:1041–1056
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Erfani J, Ebadi A, Abdollahi H, Fatahi R (2012) Genetic diversity of some pear cultivars and genotypes using simple sequence repeat (SSR) markers. PLANT MOL BIOL REP 30:1065–1072
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Fan L et al (2013) Transferability of newly developed pear SSR markers to other Rosaceae species. PLANT MOL BIOL REP 31:1271–1282
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
Hoelzel AR (2010) Looking backwards to look forwards: conservation genetics in a changing world. Conserv Genet 11:655–660
Iketani H, Yamamoto T, Katayama H, Uematsu C, Mase N, Sato Y (2010) Introgression between native and prehistorically naturalized (archaeophytic) wild pear (Pyrus spp.) populations in northern Tohoku, Northeast Japan. Conserv Genet 11:115–126
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jang JT, Tanabe K, Tamura F, Banno K (1992) Identification of Pyrus species by leaf peroxidase isozyme phenotypes. J Jpn Soc Hortic Sci (Jpn)
Janick J, Moore JN (1975) Advances in fruit breeding. Purdue University Press
Kikuchi A (1948) Horticulture of fruit trees Yokendo, Tokyo (in Japanese)
Lee SH (1948) A taxonomic survey of the oriental pears, vol 51. AMER SOC HORTICULTURAL SCIENCE, 701 NORTH SAINT ASAPH STREET, ALEXANDRIA, VA, pp 22314–21998
Lin B, Shen D (1983) Studies on the germplasmic characteristics of Pyrus by use of isozymic patterns. Acta Agriculture University Zhejiang, China 9:235–243
Liu J, Sun P, Zheng X, Potter D, Li K, Hu C, Teng Y (2013) Genetic structure and phylogeography of Pyrus pashia L. (Rosaceae) in Yunnan Province, China, revealed by chloroplast DNA analyses. Tree Genet Genomes 9:433–441. doi:10.1007/s11295-012-0564-x
Liu J, Zheng X, Potter D, Hu C, Teng Y (2012) Genetic diversity and population structure of Pyrus calleryana (Rosaceae) in Zhejiang province, China. Biochem Syst Ecol 45:69–78
Liu Q, Song Y, Liu L, Zhang M, Sun J, Zhang S, Wu J (2015) Genetic diversity and population structure of pear (Pyrus spp.) collections revealed by a set of core genome-wide SSR markers. Tree Genet Genomes 11:1–22
Mittermeier RA, Myers N, Mittermeier CG, Robles Gil P (1999) Hotspots: Earth’s biologically richest and most endangered terrestrial ecoregions. CEMEX, SA, Agrupación Sierra Madre, SC
Moritz C (1994) Applications of mitochondrial DNA analysis in conservation: a critical review. Mol Ecol 3:401–411
Newton AC, Allnutt TR, Gillies A, Lowe AJ, Ennos RA (1999) Molecular phylogeography, intraspecific variation and the conservation of tree species. Trends Ecol Evol 14:140–145
NTSYS-pc RF (2000) Numerical taxonomy and multivariate analysis system, version 2.2. Exeter Publishing, Setauket, NY, USA
Pimentel D et al (1997) Economic and environmental benefits of biodiversity. Bioscience 47:747–757
Potter D, Still SM, Grebenc T, Ballian D, Božič G, Franjiæ J, Kraigher H (2007) Phylogenetic relationships in tribe Spiraeeae (Rosaceae) inferred from nucleotide sequence data. Plant Syst Evol 266:105–118
Pritchard JK, Wen W (2003) Documentation for STRUCTURE Software: Version 2. Department of Human Genetics, University of Chicago, Chicago
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pu F, Wang Y (1963) Pomology of China pears. Shanghai Scientific and Technical Publishers, Shanghai (in Chinese)
Rehder A (1915) Synopsis of the Chinese species of Pyrus vol 50. JSTOR
Rehder A (1940) Manual of cultivated trees and shrubs
Richards CM, Volk GM, Reilley AA, Henk AD, Lockwood DR, Reeves PA, Forsline PL (2009) Genetic diversity and population structure in Malus sieversii, a wild progenitor species of domesticated apple. Tree Genet Genomes 5:339–347
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rubtsov GA (1944) Geographical distribution of the genus Pyrus and trends and factors in its evolution. Am Nat 78:358–366
Shen T (1980) Pears in China. Hortscience 15:13–17
Song Y, Fan L, Chen H, Zhang M, Ma Q, Zhang S, Wu J (2014) Identifying genetic diversity and a preliminary core collection of Pyrus pyrifolia cultivars by a genome-wide set of SSR markers SCI. HORTIC-AMSTERDAM 167:5–16
Sun Y, Du S, Yao K (1983) History of fruit culture and resources of fruit trees in China. Shanghai Sci. and Technol. Press, Shanghai
Swapna M, Sivaraju K, Sharma RK, Singh NK, Mohapatra T (2011) Single-strand conformational polymorphism of EST-SSRs: a potential tool for diversity analysis and varietal identification in sugarcane. PLANT MOL BIOL REP 29:505–513
Tautz D (1989) Hypervariabflity of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res 17:6463–6471
Teng Y, Tanabe K, Tamura F, Itai A (2002) Genetic relationships of Pyrus species and cultivars native to East Asia revealed by randomly amplified polymorphic DNA markers. J Am Soc Hortic Sci 127:262–270
Tian L, Gao Y, Cao Y, Liu F, Yang J (2012) Identification of Chinese white pear cultivars using SSR markers. GENET RESOUR CROP EV 59:317–326
Vercken E, Fontaine MC, Gladieux P, Hood ME, Jonot O, Giraud T (2010) Glacial refugia in pathogens: European genetic structure of anther smut pathogens on Silene latifolia and Silene dioica. PLoS Pathog 6:e1001229
Westwood MN, Challice JS (1978) Morphology and surface topography of pollen and anthers of Pyrus species. J Amer Soc Hort Sci 103:28–37
Wu J et al (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23:396–408
Wu J et al. (2014) High-density genetic linkage map construction and identification of fruit-related QTLs in pear using SNP and SSR markers. J Exp Bot u311
Yeh FC, Yang R, Boyle TB, Ye ZH, Mao JX (1997) POPGENE, the user-friendly shareware for population genetic analysis molecular biology and biotechnology Centre. University of Alberta, Canada, p 10
Yu T, Kuan K (1963) Taxa nava Rosacearum (1). Acta Phytotaxon Sinica 8:202–236
Yu (1979) Taxonomy of the fruit tree in China (in Chinese). China Agr. Press, Beijing
Yuanwen Teng Y, Tanabe K, Tamura F, Itai A (2001) Genetic relationships of pear cultivars in Xinjiang, China, as measured by RAPD markers. J Hortic Sci Biotechnol 76:771–779
Zhang MY, Fan L, Liu QZ, Song Y, Wei SW (2014) A Novel Set of EST-Derived SSR Markers for Pear and Cross-Species Transferability in Rosaceae. Plant Mol Biol Rep
Zheng X, Cai D, Potter D, Postman J, Liu J, Teng Y (2014) Phylogeny and evolutionary histories of Pyrus L. revealed by phylogenetic trees and networks based on data from multiple DNA sequences. Mol Phylogenet Evol 80:54–65
Zong Y, Sun P, Liu J, Yue X, Li K, Teng Y (2014) Genetic diversity and population structure of seedling populations of Pyrus pashia. PLANT MOL BIOL REP 32:644–651. doi:10.1007/s11105-013-0680-2
Acknowledgements
The work was financially supported by the National Science Foundation of China (31672111), the Earmarked Fund for China Agriculture Research System (CARS-29), the Science Foundation for Distinguished Young Scientists in Jiangsu Province (BK20150025), and The Six Talent Peaks Project in Jiangsu Province (2014-NY-025).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xue, L., Liu, Q., Qin, M. et al. Genetic variation and population structure of “Zangli” pear landraces in Tibet revealed by SSR markers. Tree Genetics & Genomes 13, 26 (2017). https://doi.org/10.1007/s11295-017-1110-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1110-7