Abstract
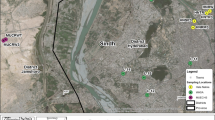
Yamuna River water in Agra city of India is contaminated with toxic pollutants, including heavy metals that cause damage to the environment and human health. At present, the direct use of river water for drinking purposes and household activities lead to the direct exposure of society to the contaminants. In this study, Yamuna River water samples were collected from three different sites in Agra city during the monsoon, summer, and winter seasons. The physico-chemical parameters were estimated along with heavy metals. In physico-chemical parameter, the values found were mostly above the permissible limits. The results water samples contain high levels of cadmium, chromium, lead, and nickel above the desirable levels in most cases. The metagenomic analysis revealed that Proteobacteria, Bacteroidetes, Verrucomicrobia, Actinobacteria, and Planctobacteria were the most abundant phyla with a relative abundance of 61%, 9.34%, 5.23%, 4.64%, and 4.3%, respectively. The Comamonadaceae, the most abundant family consists of the genera involved in hydrogen oxidation, iron reduction, degraders of polycyclic aromatic hydrocarbons, and fermentation. The presence of Pseudomonas, Nitrosomonas sp., Thauera humireducens and Dechloromonas denitrificans (decomposition of sewage and organic matter) and Pseudomonas aeruginosa indicates the presence of heavy metal degrading bacteria in water sample. Functional prediction showed the presence of genes responsible for different metabolic pathways that could help developing new bioremediation strategies. The study concludes the status of water contamination, the presence of complex microbial community and suggests the futuristic use and their role in bioremediation.








Similar content being viewed by others
Data availability
Not applicable for this manuscript.
References
Ahmed S, Akhtar N, Rahman A, Mondal NC, Khurshid S et al (2022) Evaluating groundwater pollution with emphasizing heavy metal hotspots in an urbanized alluvium watershed of Yamuna River. Northern Ind Environ Nanotechnol Monit Manag 18:100744
Alegbeleye O, Alisoltani A, Abia ALK, Awe AA, Adetunji AT, Rabiu S, Opeolu BO (2021) Investigation into the bacterial diversity of sediment samples obtained from Berg River, Western Cape, South Africa. Folia Microbiol 66:931–947
Ambasht RS (1990) Environment and pollution (An Ecological Approach). Ist ed., Students Friends and Co. Pub. Lanka Varanasi, India
Anand M, Singh L, Agarwal P, Saroj R, Taneja A (2019) Pesticides exposure through environment and risk of pre-term birth: a study from Agra city. Drug Chem Toxicol 42:471–477
APHA (1998) Standard Methods for the Examination of Water and wastewater. 19th Eds. American Public Health Association, Washington DC
Babuji P, Thirumalaisamy S, Duraisamy K, Periyasamy G (2023) Human Health Risks due to Exposure to Water Pollution: a Review. Water 15:2532. https://doi.org/10.3390/w15142532
Bai J, Li H, Zhao Y (2009) Bacterial distribution at different stations in the Northern Yellow Sea. Wei Sheng Wu Xue Bao 49:343–350
Balasaheb R, Bhalchandra W, Resham B (2015) Assessment of seasonal variations in water quality of Girna, reservoir in Nasik District (M.S.). Int J Recent Sci Res 6:6570–6573
Behera BK, Chakraborty HJ, Patra B, Rout AK, Dehury B, Das BK et al (2020) Metagenomic analysis reveals bacterial and fungal diversity and their bioremediation potential from sediments of river Ganga and Yamuna in India. Front Microbiol 11:2531. https://doi.org/10.3389/fmicb.2020.556136
Behera BK, Sahu P, Rout AK, Parida PK, Sarkar DJ, Kaushik NK et al (2021) Exploring microbiome from sediments of River Ganga using a metagenomic approach. Aquat Ecosyst Health Manag 24:12–22
Bharti M, Nagar S, Khurana H et al (2022) Metagenomic insights to understand the role of polluted river Yamuna in shaping the gut microbial communities of two invasive fish species. Arch Microbiol 204:509. https://doi.org/10.1007/s00203-022-03127-x
Bheemappa K, Nandini N, Vijaykumar M, Raghvendra M (2015) Temporal variation in water quality parameters of Bandematta Hosakere Lake-Peri urban area of Bengaluru, Karnataka, India. Int J Adv Res 3:1283–1291
BIS (Bureau of Indian Standards) (2012) “IS: 10500:2012”, Drinking Water—Specification (Second Revision), Drinking Water Sectional Committee, FAD25, India, pp 1–11
Biswas P, Vellanki BP (2021) Occurrence of emerging contaminants in highly anthropogenically influenced river Yamuna in India. Sci Total Environ 782:146741
Bose T, Haque MM, Reddy C, Mande SS (2015) COGNIZER: a framework for functional annotation of metagenomic datasets. PLoS ONE 10:0142102. https://doi.org/10.1371/journal.pone.0142102
Census of India (2011) Houselisting and housing data, Households by availability of type of latrine facility, viewed 29 May 2015, Census digital library
CPCB (2006) Water Quality Status of Yamuna River (1999 2005), Central Pollution control board, ministry of environment & forests, Assessment and development of river basin series: ADSORBS/41/2006–07
Das BK, Behera BK, Chakraborty HJ, Paria P, Gangopadhyay A, Rout AK, Rai A (2020) Metagenomic study focusing on antibiotic resistance genes from the sediments of River Yamuna. Gene 758:144951
Ding XW, Zhang JJ, Jiang GH, Zhang SH (2017) Early warning and forecasting system of water quality safety for drinking water source areas in three gorges reservoir area. China Water 9:465. https://doi.org/10.3390/w9070465
Ehrlich PR, Harte J (2015) To feed the world in 2050 will require a global revolution. Proceed Nat Acad Sci 112:14743–14744
Eknath NC (2013) The seasonal fluctuation of physico-chemical parameters of River Mula- Mutha at Pune, India and their impact on fish biodiversity. Res J Animal Veteri Fish Sci 1:11–16
Garrido Cardenas JA, Manzano-Agugliaro F (2017) The metagenomics worldwide research. Curr Genet 63:819–829. https://doi.org/10.1007/s00294-017-0693-8
Gupta D, Satpati S, Dixit A, Ranjan R (2019) Fabrication of biobeads expressing heavy metal-binding protein for removal of heavy metal from wastewater. App Microbiol Biotechnol 103:5411–5420
Harney NV, Dhamani AA, Andrew RJ (2013) Seasonal variations in the physico-chemical parameters of Pindavani Pond of Central India. Weekly Sci 1:1–8
Isaac R, Siddiqui S (2022) Application of water quality index and multivariate statistical techniques for assessment of water quality around Yamuna River in Agra Region, Uttar Pradesh, India. Water Supply 22:3399–3418. https://doi.org/10.2166/ws.2021.395
Kadiru S, Patil S, D’Souza R (2022) Effect of pesticide toxicity in aquatic environments: a recent review. Int J Fish Aquat Stud 10:13–118
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. https://doi.org/10.1093/nar/gkv1070
Khan AH, Aziz HA, Khan NA, Dhingra A, Ahmed S, Naushad M (2021) Effect of seasonal variation on the occurrences of high-risk pharmaceutical in drain-laden surface water: a risk analysis of Yamuna River. Sci Total Environ 794:148484
Khandeparker L, Kuchi N, Kale D, Anil AC (2017) Microbial community structure of surface sediments from a tropical estuarine environment using next generation sequencing. Ecol Indic 74:172–181. https://doi.org/10.1016/j.ecolind.2016.11.023
Khanna DR, Bhutiani R (2003) Limnological status of Satikund Pond at Haridwar. Ind J Environ Sci 7:131–136
Kori JA, Mahar RB, Vistro MR, Tariq H, Khan IA, Goel R (2019) Metagenomic analysis of drinking water samples collected from treatment plants of Hyderabad City and Mehran University Employees Cooperative Housing Society. Environ Sci Poll Res 26:29052–29064
Kumar M, Sharif M, Ahmed S (2020) Impact of urbanization on the river Yamuna basin. Inter J River Basin Manag 18:461–475
Lin L, Yang H, Xu X (2022) Effects of water pollution on human health and disease heterogeneity: a review. Front Environ Sci 10:880246
Mathur S, Singh D, Ranjan R (2022) Remediation of heavy metal (loid) contaminated soil through green nanotechnology. Front Sust Food Syst 6:932424
Maya K, Babu KN, Pabdmalal D, Seralathan P (2007) Hydrochemistry and dissolved nutrient flux of two small catchment rivers, south western India. J Chem Ecol 23:13–27. https://doi.org/10.1080/02757540601084029
Menzel P, Ng KL, Krogh A (2016) Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7:11257. https://doi.org/10.1038/ncomms11257
Miyashita NT (2015) Contrasting soil bacterial community structure between the phyla Acidobacteria and Proteobacteria in tropical Southeast Asian and temperate Japanese forests. Genes Genet Syst 90:61–77. https://doi.org/10.1266/ggs.90.61
NAMAMI GANGE PROGRAMME - At a Glance, September 2020. https://nmcg.nic.in/NamamiGanga.aspx.
Naz M, Turkmen M (2005) Phytoplankton biomass and species composition of lake Golbasi (Hatay-Turkey). Turk J Biol 29:49–56
Olatunji OS, Osibanjo O (2012) Determination of selected heavy metals in inland fresh water of lower River Niger drainage in North Central Nigeria. Afr J Environ Sci Technol 6:403–408. https://doi.org/10.5897/AJEST12.124
Parida PK, Behera BK, Dehury B, Rout AK, Sarkar DJ, Rai A, Mohapatra T (2022) Community structure and function of microbiomes in polluted stretches of river Yamuna in New Delhi, India, using shotgun metagenomics. Environ Sci Pol Res 29:71311–71325
Parween M, Ramanathan AL, Raju NJ (2021) Assessment of toxicity and potential health risk from persistent pesticides and heavy metals along the Delhi stretch of river Yamuna. Environ Res 202:111780
Qiu H, Gu L, Sun B, Zhang J, Zhang M, He S, Leng X (2020) Metagenomic analysis revealed that the terrestrial pollutants override the effects of seasonal variation on microbiome in river sediments. B Environ Contam Tox 105:892–898
Rafiq S, Salim R, Nielsen I (2016) Urbanization, openness, emissions, and energy intensity: a study of increasingly urbanized emerging economies. Energy Eco 56:20–28
Ravindra K, Ameena M, Monika R, Kaushik A (2003) Seasonal variation in physico-chemical characteristics of river Yamuna in Haryana and its ecological best-designated use. J Environ Monit 5:419–426. https://doi.org/10.1039/B301723K
Safwat SM, Matta ME (2021) Environmental applications of Effective Microorganisms: a review of current knowledge and recommendations for future directions. J Eng Appl Sci 68:48. https://doi.org/10.1186/s44147-021-00049-1
Saleh TA, Mustaqeem M, Khaled M (2022) Water treatment technologies in removing heavy metal ions from wastewater: a review. Environl Nanotech Monit Manage 17:100617
Samson R, Shah M, Yadav R, Sarode P, Rajput V et al (2019) Metagenomic insights to understand transient influence of Yamuna River on taxonomic and functional aspects of bacterial and archaeal communities of River Ganges. Sci Total Environ 674:288–299
Saraswat P, Singh S, Prasad M, Misra R, Rajput VD, Ranjan R (2023) Applications of bio-based nanomaterials in environment and agriculture: a review on recent progresses. Hyb Adv 4:100097
Schnetzer A, Moorthi SD, Countway PD, Gastet RJ et al (2011) Depth matters: Microbial eukaryote diversity and community structure in the eastern North Pacific revealed through environmental gene libraries. Deep Sea Res Part I 58:16–26. https://doi.org/10.1016/j.dsr.2010.10.003
Shanbehzadeh S, Dastjerdi MV, Hassanzadeh A, Kiyanizadeh T (2014) Heavy metals in water and sediment: a case study of Tembi River. J Environ Public Hlth 858720:5. https://doi.org/10.1155/2014/858720
Sharma P, Kumar S, Pandey A (2021) Bioremediated techniques for remediation of metal pollutants using metagenomics approaches: a review. J Environ Chem Engi 9:105684
Shinde SE, Pathan TS, Raut KS, Sonawane DL (2011) Studies on the physico-chemical parameters and correlation coefficient of Harsool-Savangi Dam, District Aurangabad, India. Middle-East J Sci Res 8:544–554
Singh JP, Singh S, Khanna DR (2006) Water quality of river Ganges in respect of physico-chemical and microbial characteristics at Anupshahar, district-Bulandshahar (India). Environ Conserv J 7:29–34
Singh A, Tyagi P, Ranjan R, Sushkova SN, Minkina T, Burachevskaya M, Rajput VD (2023) Bioremediation of hazardous wastes using green synthesis of nanoparticles. Processes 11:141
Sophia S, Prakash MMJ, M, (2017) Analysis and Seasonal Variation of Heavy Metals in Water and Sediment from Adyar Estuary. Environ Risk Assess Remed 1:23–28
Sustainable development goals (2015) Goal 6: Ensure access to water and sanitation for all. https://www.un.org/sustainabledevelopment/water-and-sanitation/. Accessed 12 Feb 2023
Tang J, Bu Y, Zhang XX, Huang K, He X, Ye L, Ren H (2016) Metagenomic analysis of bacterial community composition and antibiotic resistance genes in a wastewater treatment plant and its receiving surface water. Ecotoxicol Environ Safety 132:260–269
Tekin OS (2015) Levels of some heavy metals in water and sediment compared with season and some physico-chemical parameters from Antalya Bay. Ind J Geo Marine Sci 44:1393–1400
Tripathi B, Pandey R, Raghuvanshi D, Singh H, Pandey V, Shukla DN (2015) Analysis of physico-chemical parameter value and correlation coefficient of the River Ganga at Kalakankar, Pratapgarh. Asian J Biochem Pharma Res 5:180–192
Tsagaraki TM, Pree B, Leiknes Ø, Larsen A, Bratbak G, Øvreås L, Egge JK, Spanek R, Paulsen ML, Olsen Y, Vadstein O, Thingstad TF (2018) Bacterial community composition responds to changes in copepod abundance and alters ecosystemfunction in an Arctic mesocosm study. ISME J 12:2694–2705
Verma S, Singh R (2016) Physico-chemical study of Yamuna river water at Agra. J Environ Sci Comp Sci Engg Technol 5:528–533
Von Schiller D, Acuña V, Aristi I, Arroita M, Basaguren A, Bellin A, Boyero L, Butturini A, Ginebreda A, Kalogianni E, Larrañaga A, Majone B, Martínez A, Monroy S, Muñoz I, Paunović M, Pereda O, Petrovic M, Pozo J, Rodríguez Mozaz S, Rivas D, Sabater S, Sabater F, Skoulikidis N, Solagaistua L, Vardakas L, Elosegi A (2017) River ecosystem processes: a synthesis of approaches, criteria of use and sensitivity to environmental stressors. Sci Total Environ 596–597:465–480. https://doi.org/10.1016/J.SCITOTENV.2017.04.081
Wang ZH, Yang JQ, Zhang DJ, Zhou J, Zhang CD, Su XR, Li TW (2014) Composition and structure of microbial communities associated with different domestic sewage outfalls. Genet Mol Res 13:7542–7552
Wang H, Wang B, Dong W, Hu X (2016) Co-acclimation of bacterial communities under stresses of hydrocarbons with different structures. Sci Rep 6:34588. https://doi.org/10.1038/srep34588
WHO. (World Health Organisation) (2022). Guidelines for drinking-water quality: fourth edition incorporating the first and second addenda. WHO Press, World Health Organisation, Geneva, Switzerland. https://www.who.int/publications/i/item/9789240045064
Willems A (2014) The family comamonadaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The Prokaryotes. Springer, Heidelberg. https://doi.org/10.1007/978-3-642-30197-1_238
Wu H, Ge Y (2019) Excessive application of fertilizer, agricultural non-point source pollution, and farmers’ policy choice. Sustainability 11:1165
Zhang H, Huang T, Liu T (2013) Sediment enzyme activities and microbial community diversity in an oligotrophic drinking water reservoir. Eastern China Plos One 8:e78571. https://doi.org/10.1371/journal.pone.0078571
Zhou J, He Z, Yang Y, Deng Y, Tringe SG, Alvarez-Cohen L (2015) High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. Mbio 6:e02288-e12214. https://doi.org/10.1128/mBio.02288-14
Acknowledgements
Dr. Nupur Raghav would like to acknowledge the support of the Late Prof. Janendra Nath Srivastava for providing constant supervision and guidance in this study. All the authors are thankful to Xcelris Labs for extending support, wherever required, and UGC-SAP (India) for financial assistance during the research studies.
Funding
This work was supported by UGC-Botany-SAP Letter No. F.5-8/2018/DRS-I(SAP-II), India.
Author information
Authors and Affiliations
Contributions
NR and RR designed the study, NR and PS wrote the manuscript. RR. SK, AC edited the manuscript and made modifications.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Consent to participate
Not applicable to this manuscript.
Consent for publication
We give our consent and ensure that the publisher has the author’s permission to publish the research article in their Journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11274_2024_3919_MOESM1_ESM.jpg
Supplementary file1 (JPG 59 KB) The graph shows the relative abundance of water samples at the class level. From the figure, it can be inferred that Betaproteobacteria is the most abundant class followed by Gammaproteobacteria
11274_2024_3919_MOESM2_ESM.jpg
Supplementary file2 (JPG 84 KB) The graph shows the relative abundance of Water samples at the order level. From the figure, it can be inferred that Burkholderiales is the most abundant order followed by Rhodocyclales
11274_2024_3919_MOESM3_ESM.jpg
Supplementary file3 (JPG 66 KB) The graph shows the relative abundance of Water sample at the genus level. From the figure, it can be inferred that Hydrogenophaga is the most abundant genus followed by Thauera
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Raghav, N., Saraswat, P., Kumar, S. et al. Metagenomics analysis of water samples collected from the Yamuna River of Agra city, India. World J Microbiol Biotechnol 40, 113 (2024). https://doi.org/10.1007/s11274-024-03919-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-024-03919-x