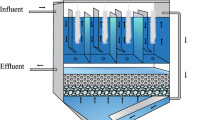
Abstract
Anaerobic dairy waste treatment requires effective control to avoid long-chain fatty acid (LCFA) inhibitory effects on anaerobic microorganisms, especially methanogens. The hybrid anaerobic baffled reactor (HABR) can provide system stability, but more needs to be done to understand how the microbial communities underpinning the HABR compartments behave and respond. Thus, this study aimed to examine the HABR’s microbial community correlating its performance when subjected to an increase in organic loading rate (OLR) during simulated dairy wastewater treatment. Besides the elevation in OLR, the system could maintain a high COD removal efficiency, nearly to 91%, and elevate the methane production to 53%. Almost all of the organic matter removal occurred mainly in C1 and C2 compartments. The genera Methanosaeta, an acetoclastic methanogen, and Methanobacterium, a hydrogenotrophic methanogen, were the HABR’s dominant species. The most representative phylum found was Bacteroidetes (12–28%), Firmicutes (3–20%), Chloroflexi (4–26%), and Proteobacteria (4–14%). Species capable of syntrophic partnership with methanogens were also identified, belonging to the family of Syntrophomonadaceae and Syntrophaceae. Microorganisms able to perform the AD process as HA73, VadinCA02, T78, Longilinea, Clostridium, and Syntrophomonas were present in the HABR.
Graphical abstract










Similar content being viewed by others
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Alves, M. M., Pereira, M. A., Sousa, D. Z., Cavaleiro, A. J., Picavet, M., Smidt, H., & Stams, A. J. M. (2009). Waste lipids to energy: How to optimize methane production from long-chain fatty acids (LCFA). Microbial Biotechnology, 2, 538–550. https://doi.org/10.1111/j.1751-7915.2009.00100.x
Amin, F. R., Khalid, H., El-mashad, H. M., Chen, C., Liu, G., & Zhang, R. (2021). Science of the total environment functions of bacteria and archaea participating in the bioconversion of organic waste for methane production. Science of the Total Environment, 763, 143007. https://doi.org/10.1016/j.scitotenv.2020.143007
Antonopoulou, G., Stamatelatou, K., Venetsaneas, N., Kornaros, M., & Lyberatos, G. (2008). Biohydrogen and methane production from cheese whey in a two-stage anaerobic process. Industrial & Engineering Chemistry, 47, 5227–5233. https://doi.org/10.1021/ie071622x
APHA, AWWA, WEF. (2012). Standard methods for examination of water and wastewater (22nd ed.). Washington, DC: American Public Health Association.
Arvin, A., Hosseini, M., Amin, M. M., Darzi, G. N., & Ghasemi, Y. (2019). A comparative study of the anaerobic baffled reactor and an integrated anaerobic baffled reactor and microbial electrolysis cell for treatment of petrochemical wastewater. Biochemical Engineering Journal, 144, 157–165. https://doi.org/10.1016/j.bej.2019.01.021
Barber, W. P., & Stuckey, D. C. (1999). The use of the anaerobic baffled reactor (ABR) for wastewater treatment: A review. Water Research, 33, 1559–1578. https://doi.org/10.1016/S0043-1354(98)00371-6
Bialek, K., Kim, J., Lee, C., Collins, G., & Flaherty, V. O. (2011). Quantitative and qualitative analyses of methanogenic community development in high-rate anaerobic bioreactors. Water Research, 5, 1298–1308. https://doi.org/10.1016/j.watres.2010.10.010
Bjorn, A., Ziels, R. M., Ejlertsson, J., Svensson, B. H., Beck, D. A. C., Stensel, H. D., Yekta, S. S., & Karlsson, A. (2016). Microbial community adaptation influences long-chain fatty acid conversion during anaerobic codigestion of fats, oils, and grease with municipal sludge. Water Research, 103, 372–382. https://doi.org/10.1016/j.watres.2016.07.043
Braga, J. K., Motteran, F., Sakamoto, I. K., & Varesche, M. B. A. (2018). Bacterial and archaeal community structure involved in biofuels production using hydrothermal- and enzymatic-pretreated sugarcane bagasse for an improvement in hydrogen and methane production. Sustainable Energy & Fuels, 2, 2644–2660. https://doi.org/10.1039/C8SE00312B
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., Fierer, N., Peña, A. G., Goodrich, J. K., Gordon, J. I., Huttley, G. A., Kelley, S. T., Knights, D., Koenig, J. E., Ley, R. E., Lozupone, C. A., McDonald, D., Muegge, B. D., Pirrung, M., Reeder, J., Sevinsky, J. R., Turnbaugh, P. J., Walters, W. A., Widmann, J., Yatsunenko, T., Zaneveld, J., & Knight, R. (2010). QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7(5), 335–336. https://doi.org/10.1038/nmeth.f.303.
Cha, I. T., Min, U. G., Kim, S. J., Yim, K. J., Roh, S. W., & Rhee, S. K. (2013). Methanomethylovorans uponensis sp. nov., a methylotrophic methanogen isolated from wetland sediment. International Journal of Molecular Microbiology, 104, 1005–1012. https://doi.org/10.1007/s10482-013-0020-4
Chen, Y., Cheng, J. J., & Creamer, K. S. (2008). Inhibition of anaerobic digestion process: A review. Bioresource Technology, 99, 4044–4064. https://doi.org/10.1016/j.biortech.2007.01.057
Chen, S., Zhang, J., & Wang, X. (2015). Effects of alkalinity sources on the stability of anaerobic digestion from food waste. Waste Management & Research, 33, 1033–1040. https://doi.org/10.1177/0734242X15602965
Chen, S., Tao, Z., Yao, F., Wu, B., He, L., Hou, K., Pi, Z., Fu, J., Yin, H., Huang, Q., Liu, Y., Wang, D., Li, X., & Yang, Q. (2020). Enhanced anaerobic co-digestion of waste activated sludge and food waste by sulfidated microscale zerovalent iron: Insights in direct interspecies electron transfer mechanism. Bioresource Technology. https://doi.org/10.1016/j.biortech.2020.123901
Damodara Kannan, A., Evans, P., & Parameswaran, P. (2020). Long-term microbial community dynamics in a pilot-scale gas sparged anaerobic membrane bioreactor treating municipal wastewater under seasonal variations. Bioresource Technology, 310123425. https://doi.org/10.1016/j.biortech.2020.123425.
Dasa, K. T., Westman, S. Y., Millati, R., Cahyanto, M. N., Taherzadeh, M. J., & Niklasson, C. (2016). Inhibitory effect of long-chain fatty acids on biogas production and the protective effect of membrane bioreactor. BioMed Research International, 2016. https://doi.org/10.1155/2016/7263974
Enzmann, F., Mayer, F., Rother, M., & Holtmann, D. (2018). Methanogens: Biochemical background and biotechnological applications. AMB Express, 8, 1–22. https://doi.org/10.1186/s13568-017-0531-x
Ferguson, R. M. W., Coulon, F., & Villa, R. (2016). Organic loading rate: A promising microbial management tool in anaerobic digestion. Water Research, 100, 348–356. https://doi.org/10.1016/j.watres.2016.05.009
Gaspar, P., Carvalho, A. L., Vinga, S., Santos, H., & Neves, A. R. (2013). From physiology to systems metabolic engineering for the production of biochemicals by lactic acid bacteria. Biotechnology. Advances, 31, 764–788. https://doi.org/10.1016/j.biotechadv.2013.03.011
Gerardi, M. H. (2003). The microbiology of anaerobic digesters. In: Wastewater microbiology series. Wiley: New Jersey.
Gorlas, A., Robert, C., Gimenez, G., Drancourt, M., & Raoult, D. (2012). Complete genome sequence of Methanomassiliicoccus luminyensis, the largest genome of a human-associated Archaea species. Journal of Bacteriology, 194, 4745. https://doi.org/10.1128/JB.00956-12
Guo, J., Peng, Y., Ni, B. J., Han, X., Fan, L., & Yuan, Z. (2015). Dissecting microbial community structure and methane-producing pathways of a full-scale anaerobic reactor digesting activated sludge from wastewater treatment by metagenomic sequencing. Microbial Cell Factories, 14, 1–11. https://doi.org/10.1186/s12934-015-0218-4
Hagen, L. H., Vivekanand, V., Linjordet, R., Pope, P. B., Eijsink, V. G. H., & Horn, S. J. (2014). Microbial community structure and dynamics during co-digestion of whey permeate and cow manure in continuous stirred tank reactor systems. Bioresource Technology, 171, 350–359. https://doi.org/10.1016/j.biortech.2014.08.095
Heuer, H., Krsek, M., Baker, P., Smalla, K., & Wellington, E. M. (1997). Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and Gel-Electrophoretic separation in denaturing gradients. Applied and Environmental Microbiology, 63, 3233–3241.
Ince, O. (1998). Performance of a two-phase anaerobic digestion system when treating dairy wastewater. Water Research, 32, 2707–2713. https://doi.org/10.1016/S0043-1354(98)00036-0
Ito, T., Yoshiguchi, K., Ariesyady, H. D., & Okabe, S. (2011). Identification of a novel acetate-utilizing bacterium belonging to Synergistes group 4 in anaerobic digester sludge. The ISME Journal, 5, 1844–1856. https://doi.org/10.1038/ismej.2011.59
Karadag, D., Köroʇlu, O. E., Ozkaya, B., & Cakmakci, M. (2015). A review on anaerobic biofilm reactors for the treatment of dairy industry wastewater. Process Biochemistry, 50, 262–271. https://doi.org/10.1016/j.procbio.2014.11.005
Karla, A., Amorim, B., Zaiat, M., & Foresti, E. (2005). Performance and stability of an anaerobic fixed bed reactor subjected to progressive increasing concentrations of influent organic matter and organic shock loads. The Journal of Environmental Management, 76, 319–325. https://doi.org/10.1016/j.jenvman.2004.02.010
Kröninger, L., Gottschling, J., & Deppenmeier, U. (2017). Growth characteristics of Methanomassiliicoccus luminyensis and expression of methyltransferase encoding genes. Archaea. https://doi.org/10.1155/2017/2756573
Kudo, Y., Nakajima, T., Miyaki, T., & Oyaizu, H. (1997). Methanogen flora of paddy soils in Japan. FEMS Microbiology Ecology, 22, 39–48.
Li, Y., & Khanal, S. K. (2017). Bioenergy principles and applications. John Wiley & Sons Inc.
Li, L., He, Q., Ma, Y., Wang, X., & Peng, X. (2015). Dynamics of microbial community in a mesophilic anaerobic digester treating food waste: Relationship between community structure and process stability. Bioresource Technology. https://doi.org/10.1016/j.biortech.2015.04.015
Li, Y., Sun, Y., Li, L., & Yuan, Z. (2018). Acclimation of acid-tolerant methanogenic propionate-utilizing culture and microbial community dissecting. Bioresource Technology, 250, 117–123. https://doi.org/10.1016/j.biortech.2017.11.034
Marzorati, M., Wittebolle, L., Boon, N., Daffonchio, D., & Verstraete, W. (2008). How to get more out of molecular fingerprints: Practical tools for microbial ecology. Environmental Microbiololy, 10, 1571–1581. https://doi.org/10.1111/j.1462-2920.2008.01572.x
McAteer, P. G., Trego, A. C., Thorn, C., Mahonya, T., Abram, F., & O’Flaherty, V. (2020). Reactor configuration influences microbial community structure during high-rate, low-temperature anaerobic treatment of dairy wastewater. Bioresource Technology. https://doi.org/10.1016/j.biortech.2020.123221
Nachaiyasit, S., & Stuckey, D. C. (1997). The effect of shock loads on the performance of an anaerobic baffled reactor (ABR). 1. Step changes in feed concentration at constant retention time. Water Research, 31, 2737–2746.
Nasr, M., Khalil, A., Abdel-kader, A., Elbarki, W., & Moustafa, M. (2014). Comparative performance between conventional and hybrid anaerobic baffled reactors for treating dairy wastewater. International Journal of Chemical and Environmental Engineering, 5, 187–194.
Palatsi, J., Illa, J., Prenafeta-Boldú, F. X., Laureni, M., Fernandez, B., Angelidaki, I., & Flotats, X. (2010). Long-chain fatty acids inhibition and adaptation process in anaerobic thermophilic digestion: Batch tests, microbial community structure and mathematical modelling. Bioresource Technology, 101, 2243–2251. https://doi.org/10.1016/j.biortech.2009.11.069
Pereira, M. A., Sousa, D. Z., Mota, M., & Alves, M. M. (2004). Mineralization of LCFA associated with anaerobic sludge: Kinetics, enhancement of methanogenic activity, and effect of VFA. Biotechnology & Bioengineering, 88, 502–511. https://doi.org/10.1002/bit.20278
Putra, A. A., Watari, T., Maki, S., Hatamoto, M., & Yamaguchi, T. (2020). Anaerobic baffled reactor to treat fishmeal wastewater with high organic content. Environmental Technology & Innovation. https://doi.org/10.1016/j.eti.2019.100586
Ramakrishnan, A., & Surampalli, R. Y. (2012). Comparative performance of UASB and anaerobic hybrid reactors for the treatment of complex phenolic wastewater. Bioresource Technology, 123, 352–359. https://doi.org/10.1016/j.biortech.2012.07.072
Salvador, A. F., Cavaleiro, A. J., Sousa, D. Z., Alves, M. M., & Pereira, M. A. (2013). Endurance of methanogenic archaea in anaerobic bioreactors treating oleate-based wastewater. Applied Microbiology and Biotechnology, 2211–2218. https://doi.org/10.1007/s00253-012-4061-9
Santos, K. A., Gomes, T. M., Rossi, F., Kushida, M. M., Del Bianchi, V. L., Ribeiro, R., Alves, M. S. M., & Tommaso, G. (2020). Water reuse: Dairy effluent treated by a hybrid anaerobic biofilm baffled reactor and its application in lettuce irrigation. Water Supply. https://doi.org/10.2166/ws.2020.276
Shen, R., Jing, Y., Feng, J., Luo, J., Yu, J., & Zhao, L. (2020). Performance of enhanced anaerobic digestion with different pyrolysis biochars and microbial communities. Bioresource Technology. https://doi.org/10.1016/j.biortech.2019.122354
Silva, S. A., Stams, A. J. M., Salvador, A. F., Cavaleiro, A. J., Sousa, D. Z., Pereira, M. A., & Alves, M. M. (2016). Toxicity of long chain fatty acids towards acetate conversion by Methanosaeta concilii and Methanosarcina mazei. Microbial Biotechnology, 9, 514–518. https://doi.org/10.1111/1751-7915.12365
Slavov, A. K. (2017). General characteristics and treatment possibilities of dairy wastewater a review. Food Technology and Biotechnology, 55, 14–28, https://doi.org/10.17113/ft.b.55.01.17.4520
Sousa, D. Z., Pereira, M. A., Alves, J. I., Smidt, H., Stams, A. J. M., & Alves, M. M. (2008). Anaerobic microbial LCFA degradation in bioreactors. Water Science and Technology, 57, 439–444. https://doi.org/10.2166/wst.2008.090
Sousa, D. Z., Pereira, M. A., Smidt, H., Stams, A. J. M., & Alves, M. M. (2007). Molecular assessment of complex microbial communities degrading long chain fatty acids in methanogenic bioreactors. FEMS Microbiology Ecology, 60, 252–265. https://doi.org/10.1111/j.1574-6941.2007.00291.x
Sousa, D. Z., Smidt, H., Alves, M. M., & Stams, A. J. M. (2009). Ecophysiology of syntrophic communities that degrade saturated and unsaturated long-chain fatty acids. FEMS Microbiology Ecology, 68, 257–272. https://doi.org/10.1111/j.1574-6941.2009.00680.x
Vanwonterghem, I., Evans, P. N., Parks, D. H., Jensen, P. D., Woodcroft, B. J., Hugenholtz, P., & Tyson, G. W. (2016). Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nature Microbiology. https://doi.org/10.1038/nmicrobiol.2016.170
Venkiteshwaran, K., Bocher, B., Maki, J., Zitomer, D. (2015). Relating anaerobic digestion microbial community and process function : Supplementary issue: Water Microbiology. Microbiol. Insights 8s2, MBI.S33593, https://doi.org/10.4137/mbi.s33593
Wainaina, S., Awasthi, M. K., & Mohammad, J. (2019). Bioengineering of anaerobic digestion for volatile fatty acids, hydrogen or methane production : A critical review. Bioengineered, 10, 437–458. https://doi.org/10.1080/21655979.2019.1673937
Westerholm, M., Schnürer, A., (2019). Microbial responses to different operating practices for biogas production systems, Anaerobic Digestion. IntechOpen, https://doi.org/10.5772/intechopen.8281519.
Wojcieszak, M., Pyzik, A., Poszytek, K., Krawczyk, P. S., Sobczak, A., Lipinski, L., Roubinek, O., Palige, J., Sklodowska, A., & Drewniak, L. (2017). Adaptation of methanogenic inocula to anaerobic digestion of maize silage. Frontiers in Microbiology, 8, 1–12. https://doi.org/10.3389/fmicb.2017.01881
Wu, G., Li, Z., Huang, Y., Zan, F., Dai, J., Yao, J., Yang, B., Chen, G., & Lei, L. (2020). Electrochemically assisted sulfate reduction autotrophic denitrification nitrification integrated (e-SANI®) process for high-strength ammonium industrial wastewater treatment. Chemical Engineering Journal. https://doi.org/10.1016/j.cej.2019.122707
Xia, D., Yi, X., Lu, Y., Huang, W., Xie, Y., Ye, H., et al. (2019). Dissimilatory iron and sulfate reduction by native microbial communities using lactate and citrate as carbon sources and electron donors. Ecotoxicology and Environmental Safety, 174, 524–531. https://doi.org/10.1016/j.ecoenv.2019.03.005
Xu, F., Li, Y., Ge, X., Yang, L., & Li, Y. (2018). Anaerobic digestion of food waste – Challenges and opportunities. Bioresource Technology, 247, 1047–1058. https://doi.org/10.1016/j.biortech.2017.09.020
Yin Q, He K, Liu A, Wu G., (2017). Enhanced system performance by dosing ferroferric oxide during the anaerobic treatment of tryptone-based high-strength wastewater, 3929–39. https://doi.org/10.1007/s00253-017-8194-8.
Zhender, A. J. B., Huser, B. A., Brock, T. D., & Wuhrmann, K. (1980). Characterization of an acetate-decarboxylating, non-hydrogen-oxidizing methane bacterium. Archives of Microbiology, 124, 1–11. https://doi.org/10.1007/BF00407022
Zhou, Z., Tao, Y., Zhang, S., Xiao, Y., & Meng, F. (2019). Size-dependent microbial diversity of sub-visible particles in a submerged anaerobic membrane bioreactor (SAnMBR ): Implications for membrane fouling. Water Research, 159, 20–29. https://doi.org/10.1016/j.watres.2019.04.050
Ziels, R. M., Beck, D. A. C., Martí, M., Gough, H. L., Stensel, H. D., & Svensson, B. H. (2015). Monitoring the dynamics of syntrophic β-oxidizing bacteria during anaerobic degradation of oleic acid by quantitative PCR. FEMS Microbiology Ecology, 91(4). https://doi.org/10.1093/femsec/fiv028.
Zuo, X., Yuan, H., Wachemo, A. C., Wang, X., Zhang, L., Li, J., Wen, H., Wang, J., & Li, X. (2020). The relationships among sCOD, VFAs, microbial community, and biogas production during anaerobic digestion of rice straw pretreated with ammonia. The Chinese Journal of Chemical Engineering, 28, 286–292. https://doi.org/10.1016/j.cjche.2019.07.015
Funding
The authors would like to acknowledge financial support from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – CAPES Finance Code 001 and process no. 23038.009805/2012–39.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Giordani, A., Brucha, G., Santos, K.A. et al. Performance and Microbial Community Analysis in an Anaerobic Hybrid Baffled Reactor Treating Dairy Wastewater. Water Air Soil Pollut 232, 403 (2021). https://doi.org/10.1007/s11270-021-05348-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-021-05348-0