Abstract
We aimed to evaluate the possibility the nuclear transformation of lettuce and tobacco to produce recombinant anti-HIV microbicide griffithsin under impact of Zera signal peptide. For this purpose, the codon optimized GRFT fused with KDEL retention signal was used with and without the Zera (γ-zein ER-accumulating domain) signal peptide. Integration of GRFT into the nuclear genome of lettuce and tobacco transgenic lines was confirmed by polymerase chain reaction (PCR) and Southern blot analysis. Subsequent reverse transcription-quantitative PCR (RT-qPCR) experiments showed highly divergent GRFT expression patterns, inherent to the applied transformation procedure. The recombinant GRFT was successfully detected by means of western blot and quantified by ELIZA. According to ELIZA results, fusion of GRFT with Zera signal peptide resulted in higher accumulation of the recombinant protein in both species once compared with transgenic line without signal peptide. Lettuce showed higher transgene transcripts and accumulated more the recombinant protein of interest (up to 8.942 µg/100 mg) than tobacco. Both lettuce- and tobacco-derived GRFT (GRFTL and GRFTT, respectively) captured gp120 in a way comparable to E. coli expressed GRFT (GRFTE). Our results suggest that lettuce as a leafy vegetable crop and tobacco as a model plant in transgenic research studies can be used as suitable candidate hosts for the production of recombinant GRFT, augmented by recruitment of plant optimized codon compositions and suitable signal peptide.
Similar content being viewed by others
Introduction
Human Immunodeficiency Virus (HIV) continues to pose a global threat, with about 36.7 million people infected worldwide (WHO 2016). Beside the search for efficient vaccines and therapeutic approaches, prevention of viral entry on the site of infection is a promising measure to reduce the spread of HIV. The virus crosses mucosal epithelium and infects T helper cells bearing CD4 receptors (Heron and Elahi 2017). Viral envelope glycoproteins gp120 and gp41, decorated with mannose-rich glycans, facilitate binding of the HIV particles to their target CD4 molecules (Cao et al. 2017). An assortment of lectins, or carbohydrate binding agents (CBAs), targeting the aforementioned mannose-rich moieties, block the HIV-CD4 interaction and, consequently, cell to cell transmission of the virus (Mitchell et al. 2017). Among lectins that have been used to harness HIV, griffithsin (GRFT) is the most potent agent. The protein, first extracted from the red algae Griffithsia sp., possesses six carbohydrate binding sites—a feature most probably accounting for its extraordinary properties (O’Keefe et al. 2009). GRFT might prove an ideal ingredient of a topical HIV microbicide due to its lack of toxicity, T-cell activation, natural lectin attributes, environmental stability (resistance to a broad range of pH values and high temperatures), and stability in cervicovaginal lavage fluid (established in macaques) coupled with potent antiviral activity (25,000-fold more effective than other lectins) (Mori et al. 2005; O’Keefe et al. 2009). However, different expression platforms for production of this potential topical drug ingredient are in progress in plants (Fuqua et al. 2015b; Hahn et al. 2015; Vamvaka et al. 2016), for the continuous clinical progress of GRFT, an efficient and inexpensive production platform is required in parallel to its’s clinical development (Fuqua et al. 2015a).
Recombinant GRFT was successfully produced in Escherichia coli (70% of protein in soluble fraction) (Giomarelli et al. 2006) and, in multigram quantities, in Nicotiana benthamiana plants transduced with a tobacco mosaic virus (TMV) vector (O’Keefe et al. 2009). Although these platforms provide sufficient amounts of recombinant GRFT, the relatively high capital investment in bacterial fermenters or the tedious inoculation procedure for transient plant-based expression constitute their respective drawbacks (Dirisala et al. 2016). Furthermore, the enumerated issues might prove insurmountable hurdles for GRFT production in developing countries and, therefore, limit its preventive usage in HIV-endemic areas.
Stably transformed plants have been shown to provide a versatile alternative to conventional pharmaceutical protein production systems, such as bacteria, yeast, and mammalian cell cultures (Habibi et al. 2017). Depending on the product yield, recombinant proteins can be efficiently generated in planta at 2–10% cost of microbial fermentation systems and 0.1% cost of mammalian cell cultures, without the risk of human or zoonotic pathogen transmission (Buyel et al. 2017). Although the production levels are lower than those obtained through application of transient expression systems, once established, stable transgenic lines, grown and propagated employing simple agricultural practices, provide a sustainable source of recombinant proteins—a viable and cost-effective production platform (Mortimer et al. 2015). In this regard, Hahn et al. (2015) could produce recombinant GRFT in endosperm of transgenic rice plants (Oryza sativa). They also introduced a one-step purification method to develop large-scale process and facilitate an inexpensive downstream processing platform.
In the recent years, different strategies have been developed to avoid recombinant proteins degradation and to improve their expression and accumulation in prokaryotic and eukaryotic hosts. Among these approaches, targeting of expressed proteins to special cellular compartments has attracted a special attention. In this regard, plants offer a multitude of localization compartments, both on cellular and sub-cellular level, for recombinant protein accumulation and sequestration (safeguarding against the detrimental influence of, e.g., specific proteases) (Warzecha 2008). One of the recent developments, the Zera leader peptide, was shown to enhance recombinant protein accumulation (Conley et al. 2011; Llop-Tous et al. 2011; Torrent et al. 2009). The 112 amino acids of Zera, the N-terminal domain of γ-zein (a maize storage protein), comprise a 19-amino acid signal peptide, a proline-rich repeat region containing eight units of the hexapeptide PPPVHL and a proline-X sequence encompassing four cysteines; the enumerated features induce protein body formation, not only in plants but also in filamentous fungi, as well as mammalian and insect cells, further facilitating simple and inexpensive downstream processing (Conley et al. 2011; Saberianfar et al. 2016). Llop-Tous et al. (2011) have successfully generated functional insoluble aggregates of xylanase enzyme fused to Zera and KDEL in tobacco plants proving protein body formation and simple density-based downstream process.
In a previous study, we developed a versatile modular cloning toolbox facilitating chloroplast transformation, highlighting its efficiency through successful GRFT expression in tobacco (Vafaee et al. 2014). Here, we aimed at (i) the evaluation of transgenic Lactuca sativa cv. TN-96-39 and Nicotiana tabacum cv. Samsun plants as stable sources of recombinant GRFT (ii) to explore if the presence of N-terminal ZERA signal peptide can improve KDEL-fused GRFT expression and accumulation.
Materials and methods
Codon optimization and vector construction
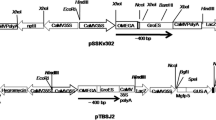
Unsuitable restriction sites within the GRFT gene sequence (NCBI, AY744143.1) were determined by means of the New England BioLabs online tool, NEBcutter and removed in silico. To ensure highest GRFT expression, based on both lettuce and tobacco codon usage frequency (Kazusa DNA Research Institute 2016), some parts of the gene sequence were modified based on plant codon preferences (Fig. 1). The resulting sequence was further customized to ensure high degree of codon diversity, desirable base composition, and absence of sequences that might interfere with transcription or translation processes. The synthesized GRFT sequence (ShineGene Molecular Biotech Inc., China) tailed with KDEL targeting signal was cloned into the binary vector pBI121 (Clontech Laboratories, USA), harboring the cauliflower mosaic virus (CaMV) 35S promoter, neomycin phosphotransferase gene (nptII) as a selectable marker conferring kanamycin resistance, and nopaline synthase (nos) terminator, resulting in the generation of the pB121KD-gR construct. For the set-up of the pB121ZRKD-gR construct, the Zera signal peptide (NCBI, AF371261.1), initially amplified from maize seedling genomic DNA using Zera-F: 5ʹ-AACTCTAGATAAACAATGAGGGTGTTGCTCGTTGCCC and Zera-R: 5ʹ-ATGAATTCGGATCCCTGGCACGGGCTTGGATGCGG, cloned into pGEPA-LTB harboring GRFT-KDEL fusion and was finally removed from the parent construct by XbaI–BamHI double digestion and introduced to the pBI121 vector (Fig. 2).
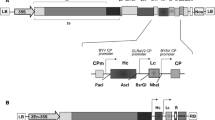
The schematic presentation of codon optimized Griffithsin (GRFT) expressed in lettuce and tobacco plants with encoding amino acids [9] aligned with original wild type sequence from Griffithsia sp. Optimized codons are indicated in the pie chart with red letters and the changes to the original nucleotides are shown by lower cases. A total 71 out of 121 codons (about 58.5%) were substituted. Location of the Kozak sequence, thrombin cleavage site, polyhistidine-tag, and KDEL targeting signal are indicated. ‘‘ATG’’ and “TAA” which denote the ‘‘Start’’ and termination codons, respectively are shown by blue color. (Color figure online)
Schematic representation of expression cassettes anchored within vectors used for lettuce and tobacco transformation. a pB121KD-gR, GUS was replaced with GRFT and fused with the KDEL targeting signal sequence. b pB121ZRKD-gR, GRFT combined with the Zera signal peptide. RB and LB, right and left borders; Pnos, nopaline synthase promoter; nptII, neomycin phosphotransferase gene (selectable marker); Tnos, nopaline synthase terminator; PCaMV35S, cauliflower mosaic virus 35S promoter
Plant material and transformation
For regeneration of lettuce (L. sativa cv. TN-96-39), according to previous regeneration protocols (Curtis 2005; Pniewski et al. 2017), seven media were used for shoot regeneration, elongation, and root formation (Table 1). In case of tobacco (N. tabacum cv. Samsun), regeneration and transformation procedures were based on the Clemente (2005) protocol. For transformation, axenically sterilized lettuce seeds were cultured on ½ MS (Murashige and Skoog 1962) hormone-free medium and incubated at 25 ± 1 °C with a 16 h photoperiod (radiation, 22 µmol m−2s−1). Then, four days old cotyledons were placed upside down on pre-culture medium containing 0.2 mg/L benzyladenine (BA) and 0.05 mg/L naphthalene acetic acid (NAA) for 48 h.
Wounded tobacco leaf fragments and lettuce cotyledons were inoculated with A. tumefaciens LBA4404 cells (in half strength MS medium) harboring the aforementioned test vectors (OD600 of the cell suspensions, 0.6) for ten and two min, respectively, and incubated on co-cultivation medium (antibiotic-free MS containing the pre-culture medium hormones) for 24 h. The explants were transferred to the selective medium (MS containing 0.2 mg/L BA, 0.05 mg/L NAA, 300 mg/L cefotaxime, and 10 and 100 mg/L kanamycin, for lettuce and tobacco respectively). The selection medium was changed every 14 days until kanamycin-resistant shoots could be transferred to the rooting medium (hormone-free half strength MS supplemented with 50 mg/L kanamycin and 300 mg/L cefotaxime). After acclimatization, kanamycin-resistant plantlets were transferred to the greenhouse.
PCR and Southern blot analyses
After extracting total genomic DNA from ten young regenerated shoots of both lettuce and tobacco, GRFT specific primers (GRFT-F: 5ʹ-GGTGGTTCTCCTTTCTCTGG and GRFT-R: 5ʹ-AAGATAATCACCAGCACTACCG) were used for the generation of 303 bp gene amplicons in lettuce-derived samples, while the forward primer specific to the CaMV 35S promoter sequence (CaMV35S-F: 5ʹ-CTTCAAAGCAAGTGGATTGATGTGATATCTCC) in combination with GRFT-R were employed for the amplification of the corresponding 1110 bp fragments from tobacco DNA isolates. For Southern blot analysis, 20 µg of genomic DNA was digested with EcoRI, subjected to electrophoresis on a 0.9% agarose gel at 25 V for 12 h, and transferred to the positively charged Amersham Hybond-N + nylon membrane (GE Healthcare Life Sciences, UK) by capillary blotting. A 303 bp DIG-labeled GRFT probe was amplified with the PCR-DIG Probe Synthesis Kit (Roche Diagnostics, Germany). After hybridization at 42 °C for 16 h, the membrane was washed with 2 × SSC buffer (0.3 M NaCl; 0.03M Na-Citrate; pH7.0) for 15 min and 0.5 × SSC buffer for 30 min at room temperature. Probe-target hybrids were detected with an alkaline phosphatase conjugated antibody, using NBT/BCIP as a substrate (Roche Diagnostics, Germany). Imaging was performed by means of the GS-800 Imaging Densitometer (Bio-Rad, USA).
RT-PCR and real-time PCR analyses
Total RNA from PCR-positive plants of both species (lettuce and tobacco) and each constructs (pB121KD-gR and pB121ZRKD-gR, 6 plants each) was extracted with bioZOL™ (bioWORLD, USA) according to the supplier’s manual. Reverse transcription of the DNase (Thermo Fisher Scientific, Germany) treated RNA was performed with the RevertAid Reverse Transcriptase (Thermo Fisher Scientific, Germany) using an Oligo (dT)-25 primer at 40 °C for 1 h. RT-PCR was then carried out with GRFT-RT-F: 5′-GGTGGTAACCTTTCTCCTACTTTCAC and GRFT-RT-R: 5′-CCACCATAAGGACCGAATCTTC specific primers. β-Actin primers specific for lettuce (LsACT-F: 5′-CATCTATGATTGGAATGGAAGCTG and LsACT-R: 5′-TCATCCGGTCAGCAATGC) and tobacco (NtACT-F: 5′-TGAAGGTTACGCCCTTCCTC and NtACT-R: 5′-GCGGACAATTTCCCGTTC) were added to the RT-PCR reactions to provide an internal control for RNA quantity. Real-time PCR (qPCR) was performed using the Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific, USA) according to the supplier’s manual. The conditions were: 95 °C for 10 min, followed by 40 cycles of amplification (95 °C for 15 s and 60 °C for 1 min). Additionally, a 150 bp fragment of the lettuce β-actin gene (NCBI, AB290314) and its 127 bp tobacco counterpart (NCBI, CS255217.1) were synthesized, paralleling the GRFT amplification. The aforementioned (RT) primers were used for all qPCR experiments. The Ct value calculations were performed with the use of the REST software (Pfaffl et al. 2002) and the relative expression levels of GRFT were represented as \({{\text{2}}^ - }^{{\Delta \Delta {C_{\text{t}}}}}\) values, according to Livak and Schmittgen (2001), with the transcript level characteristic of the lowest-expressing transgenic line (TLK3) determined as the baseline value.
Enzyme-linked immunosorbent assay
Total amount of soluble proteins (TSP) was extracted by grinding 100 mg leaf samples in liquid nitrogen to a fine powder and resuspending in 2 mL of protein extraction buffer containing 50 mM Tris–HCl (pH 8.0), 0.5 M NaCl, 2 mM EDTA, and 1 mM dithiothreitol (DTT). The samples were centrifuged at 12,000×g for 25 min at 4 °C and protein concentration was established by means of the Bradford (1976) assay. Accumulation of the recombinant GRFT was determined by enzyme-linked immunosorbent assay (ELISA). First, the test 96-well polystyrene microplates (Costar, USA) were coated with 50 µL of TSP extracts derived from the transgenic lines exhibiting highest GRFT expression levels, as determined by qPCR, and incubated at 4 °C overnight. In parallel, serial dilutions of in-house E. coli-generated GRFT (data not shown) were dispensed into the plate wells to facilitate the delineation of a standard curve for the estimation of the target protein concentration. After washing with 1% (w/v) fat-free powdered milk in PBST for 1 h at 25 °C, the plates were incubated at 37 °C for 1 h with an in-house generated mouse polyclonal anti-GRFT antibody (unpublished data). The horseradish peroxidase-conjugated anti-mouse IgG (Pasteur Institute, Iran) was added as a secondary antibody and the plates were incubated at room temperature for 1 h. The TMB peroxidase substrate (Merck, Germany) was used to visualize antibody binding. The reaction was stopped with 1 N HCl and the optical density at 450 nm (OD450) was measured using the 2100 UV Spectrophotometer (UNICO, USA). The obtained absorbance values were then compared with the standard curve for the quantification of GRFT accumulation. All assays were performed in triplicate and the resulting data statistically analyzed as described above.
GRFT purification and western blot analysis
To confirm stability and heritability of GRFT transgene in lettuce and tobacco, one transgenic lettuce and tobacco lines from T3 generation along with E. coil (pET21 strain) harboring GRFT gene were selected for protein purification and western blot analysis. For purification of GRFT, a gravity-flow purification of hexahistidine-tagged protein was carried out using Ni–NTA agarose resin (Qiagen, Hilden, Germany) under native condition. Equilibration of resin was done based on the manufacturer’s instructions. Then, 50% Ni–NTA slurry was mixed with 5 ml protein supernatant (obtained from E. coil, lettuce and tobacco) and was gently shaken on a rotary shaker (200 rpm on) at 4 °C for 1 h. It was then loaded on a chromatography column allowed to drain by gravity at flow rates of 1 ml/min. Weakly bound proteins were eliminated by adding washing buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole pH 8.0) to the columns. The recombinant GRFT was consequently eluted from columns using elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole pH 8.0 and 20% glycerol). Fractions containing recombinant GRFT were then dialyzed against elution buffer and reacted with thrombin for 12 h at 4 °C and finally stored at − 80 C. Elimination of 6xHis-tag thrombin cleavage site from C-terminal of purified recombinant GRFT and subsequent removal of biotinylated thrombin mediated by Streptavidin Agarose was carried out using Thrombin Cleavage Capture Kit (Merck) based on the protocol provided by manufacturer. Western blot analysis was performed using the primary in-house generated mouse polyclonal anti-GRFT antibody. Purified GRFT from E. coli, lettuce and tobacco plants were used for western blot analysis. For this, electroblotting was done using a semi-dry blotting apparatus. Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane at 100 V for 60 min. After adding in-house generated polyclonal anti-GRFT antibody, washing with PBST buffer and incubating with horseradish peroxidase-conjugated secondary antibody for 60 min, the antigen–antibody complexes were visualized using a chemiluminescent kit (ECL, Amersham) according to the manufacturer’s instructions.
gp120 binding assay
ELISA was employed to study the binding of recombinant GRFT to gp120. For this purpose, GRFT expressed in E. coil as well as GRFT produced in selected tobacco and lettuce plants from western blot experiment was used for gp120 neutralization analysis. In brief, the gp120 protein diluted in PBS (Sino Biological) was bound to a Nunc MaxiSorp® 96-well plate (Thermo Fischer Scientific) at 100 ng per well by incubation at room temperature for 2 h. The wells were then rinsed three times with TPBS (1% PBS and 0.1% Tween) and blocked with BSA (1% Bovine Serum Albumin). After being washed again thrice with TPBS, 50 ng of GRFT was added to both of wells of the BSA control plates and wells treated with gp120 followed by addition of 1:1000 dilution of the anti-GRFT mouse polyclonal antibody solution. The bound GRFT was determined by adding rabbit anti-mouse antibodies conjugated to horseradish peroxidase (Pasteur Institute, Iran). The plates were then washed again with TPBS, and 3,3,5,5-etramethylbenzidine peroxidase substrate solution (AppliChem) was added to the plates. The reaction was finally stopped by the inclusion of 50 l/well of 2 M 2 M sulfuric acid, and absorbance was measured at 450 nm. All assays were performed in triplicate.
Statistical analyses
The obtained Ct values of GRFT gene expression were subjected to the analysis of variance (ANOVA) and the means were compared by Duncan’s multiple-range test (DMRT) at the 5% probability level. Two sample analyses was performed by t-test using means of mean relative gene expression, GRFT accumulation and regeneration efficiency. All statistical analyses were performed using SPSS (V. 16.0).
Results
The synthetic GRFT gene was optimized based on the codon usage of L. sativa and N. tobacum without modifying the amino acid sequence. In total, 71 codons from 121 codons of GRFT coding sequence were optimized where synthetic gene sequence was only 56% identical to the wildtype gene sequence (Fig. 1). Furthermore, rare codons, direct sequence repeats, cryptic splice sites and endoribonuclease cleavage sites were eliminated. Figure 2 represents the expression cassettes anchored within vectors used for lettuce and tobacco transformation.
To establish an effective leaf-based regeneration and transformation system for lettuce, we used seven different media (Table 1). Among these, SF2 (supplemented with 0.2 mg/L of BA and 0.05 mg/L of NAA) provided for the highest direct shoot regeneration rate (Table 1; Fig. 3b). In contrast, the highest number of indirect regeneration events from calli took place with SF1 containing 0.1 mg/L BA and 0.1 mg/L NAA (Table 1; Fig. 3e). Although all three media employed for root induction proved efficient, shoots grown on hormone-free ½ MS medium showed most effective root formation (Table 1; Fig. 3h). Once the lettuce explants were transformed with the pB121KD-gR and pB121ZRKD-gR constructs, 54 putative kanamycin-resistant transformants were regenerated and 20 PCR-positive transformant lines with highest transgene expression (data not shown) were rooted and transferred to the greenhouse for flowering and seed production. Of 336 starting explants of each investigated host species, 162 green kanamycin-resistant shoots were obtained in case of tobacco, while the lettuce-specific quotient was 53 (Table 2).
Regeneration and genetic transformation of L. sativa cv. TN-96-39 and N. tabacum cv. Samsun (left and right sub-panels, respectively). a Direct shoot regeneration from lettuce cotyledons and tobacco leaf explants on MS medium (containing kanamycin, cefotaxime, BA, and NAA), 18 days after transformation (without callus formation). b Regenerated kanamycin-resistant shoots, 4 weeks after transformation. c Non-transformed lettuce cotyledons and tobacco leaf explants on MS containing kanamycin. d Kanamycin-resistant calli formed after 3 weeks on MS medium (containing kanamycin, BA, and NAA). e Kanamycin-resistant shoots regenerated from calli shown in d. f Bleaching of non-transformed shoots under pressure of kanamycin. g Elongated shoots on hormone-free MS medium containing kanamycin. h Rooting on hormone-free ½ MS medium. i Fully acclimatized T0 lettuce and tobacco transgenic lines in the greenhouse
Based on the results of PCR, a 303 bp amplicons representative of GRFT gene was successfully amplified in the putative lettuce transgenic lines. For tobacco, PCR with the CaMV 35S promoter (forward) and the GRFT (reverse) primers resulted in the amplification of a 1110 bp fragment in all transgenic lines, whereas no corresponding PCR products were detected in the wild type (WT) plants (Fig. 4a). Southern blot analysis further corroborated genomic integration of GRFT and facilitated determination of the number of transgene insertions. The generated tobacco and lettuce lines exhibited two to three GRFT insertion sites, while WT plants showed no signal (Fig. 4c).
a PCR amplification of a 303 bp GRFT fragment from the genomic DNA of transgenic lettuce and a 1110 bp fragment (CaMV 35S promoter sequence plus GRFT) from the DNA of transformed tobacco. b Expression analysis by RT-PCR; complementary DNA fragments of GRFT (134 bp) and β-actin (150 and 127 bp, internal control). c Southern blot analysis of lettuce and tobacco transgenic lines using a DIG-labelled GRFT probe. TLZ and TL, transgenic lettuce plants carrying GRFT-KDEL fusion with and without Zera signal peptide, respectively; TT and TTZ, their respective tobacco counterparts; WT, non-transformed lettuce plant; W, water sample as a control for the amplification of the β-actin fragment; M, 1 kb GeneRuler DNA Ladder (Thermo Fisher Scientific, Germany); PC, positive control (pB121KD-gR containing GRFT)
Amplification of a 134 bp fragment from the cDNA of both species with the GRFT specific primers confirmed successful transcription of the gene of interest. For optimization of the subsequent real-time PCR experiments, the 150 and 127 bp β-actin fragments were amplified as reference house-keeping genes for lettuce and tobacco, respectively (Fig. 4b). The relative GRFT transcript levels, as illustrated by the\({{\text{2}}^ - }^{{\Delta \Delta {C_{\text{t}}}}}\)\({{\text{2}}^ - }^{{\Delta \Delta C_{{\text{t}}}^{{}}}}\) \({{\text{2}}^{ - \Delta \Delta }}^{{{C_{\text{t}}}}}\)\(({{\text{2}}^ - }^{{\Delta \Delta {C_{\text{t}}}}})\) values, were highest for the transgenic tobacco TT1 and the transgenic lettuce TLZ1, with 26.071 and 20.398 mean fold change in gene expression in the respective lines, as compared to the TL3 lowest-expressor (Fig. 4). The mean relative transcript level values of studied transgenic lines revealed no significant difference between lettuce and tobacco. The immunoassay results showed that the levels of recombinant GRFT from transgenic events ranged from 0.295 µg/100 mg fresh leaf mass in TL3 to 8.942 µg/100 mg in TLZ4 (Fig. 5a), which corresponded to 0.011 and 0.311% of TSP content in the respective lines. In general, transgenic lettuce accumulated significantly higher amounts of the recombinant protein of interest than tobacco. Moreover, both lettuce and tobacco transformants characterized by the N-terminal fusion with the Zera signal peptide showed a significant increase in GRFT accumulation as compared to those without signal peptide (Table 3). The subsequent western blot analysis using anti-GRFT polyclonal antibody of the soluble protein extracts from E. coli harboring GRFT transgene (GRFTE) led to the detection of the expected molecular size of 13 kDa while two bands of 13 and 27 kDa were visualized from transgenic lettuce (GRFTL) and tobacco (GRFTT) lines (Fig. 5b). The extra band at 27 kDa implying the possible presence of dimer form of the GRFT protein. In accordance with ELIZA and western blot analyses, SDS-PAGE analysis of purified GRFTE, GRFTL and GRFTT using Coomassie blue staining further confirmed the existence of the correctly folded and soluble recombinant protein at 13 kDa molecular which was consistent with the expectation (Fig. 5c).
a Quantification of GRFT accumulation by means of ELISA. Four plants of each species with and without Zera signal peptide were analyzed. TL and TLZ, transgenic lettuce plants carrying GRFT-KDEL with and without Zera signal peptide GRFT fusions, respectively; TT and TTZ, their respective tobacco counterparts; WT, non-transformed lettuce plant. Relative values are represented as mean ± standard error of three independent experiments with three replicates of transgenic plants developed with each vector. b Western blot analysis of total soluble protein extracted from E. coil (GRFTE) as positive control, lettuce (GRFTL) and tobacco (GRFTL) using anti-GRFT polyclonal antibody. c SDS-PAGE analysis of GRFT purified with Ni–NTA agarose resin. E.coli-L: flowthrough obtained from soluble E. coli lysate, E.coli-L: flowthrough obtained after adding binding buffer, GRFTE: final purified GRFT from E. coli, GRFTL and GRFTP: final purified GRFT from lettuce and tobacco leaf extracts, respectively, M, Pre-stained Protein Marker (Thermo Fisher Scientific)
The purified GRFT from T3 plant of transgenic lettuce and tobacco was evaluated by ELISA for in vitro binding activity against HIV-1 envelope glycoprotein gp120, employing E. coli–expressed GRFT (GRFTE) as a positive control. As represented in Fig. 6, GRFTL and GRFTT captured gp120 in a way comparable to E. coli expressed GRFT (GRFTE), thereby suggesting that plant-expressed GRFT had essentially the common oligosaccharide and dose-dependent gp120 binding activity nearly identical to the recombinant GRFT expressed in E. coli. In this regard, the highest OD450 was obtained with GRFT purified from lettuce (GRFTL) at 100 ng/well concentration.
Comparison of gp120 binding of GRFT purified from E. coli with GRFT purified from lettuce and tobacco determined by ELISA. Values are represented as the average ± SE of three wells. Plates coated with HIV-1 gp120 were treated with different concentrations of purified GRFT from E. coli (GRFTE), lettuce (GRFTL) and tobacco (GRFTT) and binding activity was measured using a primary mouse anti-GRFT polyclonal Abs and a secondary HRP-labeled antibody. Studied concentrations were 0.01, 0.1, 1, 10, 100 and 1000 ng purified GRFT per well. OD optical density at 450 nm
Discussion
In this study, we demonstrated the successful expression of a differentially fused, codon-optimized GRFT gene in lettuce and tobacco leaf to establish a versatile platform for the production of high value pharmaceuticals. Expression of GRFT-KDEL fusion was achieved with and without Zera signal peptide implying that subcellular targeting could improve the final yield of recombinant GRFT protein.
Routine tissue culture protocols and well-established genetic transformation methods render tobacco the most prominent plant host in molecular farming (Clemente 2005; Moustafa et al. 2016). However, high alkaloid content prevents direct use of tobacco leaf extracts and might pose risk even after purification (Appaiahgari et al. 2017). Lettuce, in turn, is a leafy vegetable affording high biomass and considerable regeneration capacity that validate the plant as a promising candidate for transformation endeavors especially to develop edible vaccine. In this regard, and in accord with previous studies (Darqui et al. 2018; Liu et al. 2012; Pniewski et al. 2017), we obtained a large number of initial transformants from cotyledons of the TN-96-39 lettuce cultivar. Due to the highest observed direct shoot regeneration rate, we selected SF2 medium (MS supplemented with 0.05 mg/L of NAA and 0.2 mg/L of BA) for further lettuce transformation efforts.
Transgene introduction into the plant genome is a complicated process involving different steps of recombination (Gelvin 2017). Scaffold-attachment regions (SARs) or matrix-associated regions (MARs) are 300–500 bp AT-rich sequences encompassing DNA-destabilizing (unwinding) elements (Singh et al. 2016) that enhance transgene integration in various host systems and ameliorate transgene silencing (Zhao et al. 2017). It has been demonstrated that high genetic transformation capacity of tobacco is mainly associated with the presence of strong SAR regions in its nuclear genome (Allen et al. 2001). The aforementioned higher transformation efficiency of tobacco (vs. lettuce) observed in the present study can thus be traced to its chromosomal make-up featuring multiple SAR sequences. Moreover, variable expression levels of transgenes can be attributed to different factors, such as gene copy number variation and position effect (Gelvin 2017; Rajeev Kumar et al. 2015; Tang et al. 2007). Therefore, the pronounced differences in the obtained GRFT expression values between the individual transgenic lettuce and tobacco lines (Fig. 7) might probably imply the influence of position effect (Dolgova et al. 2015), involving illegitimate T-DNA integration into genome regions which could be near to or far from transcriptional activating elements or enhancers, resulting in unpredictable transgene expression patterns—a common phenomenon among nuclear transgenic events spurred through application of the Agrobacterium-based plant transformation solutions (Habibi et al. 2017; Rajeev Kumar et al. 2015).
Expression analysis of GRFT at transcript level relative to actin as a reference gene. Four PCR-positive lettuce and tobacco plants harboring GRFT with and without Zera signal peptide were analyzed. TL and TLZ, transgenic lettuce plants carrying GRFT with and without Zera signal peptide, respectively; TT and TTZ, their respective tobacco counterparts; WT, non-transformed lettuce plant. Data are represented as mean values ± standard error of three independent replicates of transgenic plants developed with each vector
Real-time PCR, coupled with reverse transcription (RT-qPCR), was employed as a tool for monitoring and quantification of transgene expression (Schmidt and Parott 2001; Zhang et al. 2015). Our investigation showed that both lettuce and tobacco possess high capacity for GRFT transcription under control of the 35S promoter (Fig. 4). However, probably due to RNA turn-over and posttranscriptional modification(Elvira-Matelot and Martínez 2017), accumulation of the recombinant protein did not directly correlate with the transcript levels established herein. Despite significantly higher regeneration efficiency (48.21%) and relatively higher transgene expression (8.561 mean relative expression) characteristic of tobacco, the mean recombinant GRFT accumulation values were higher in transgenic lettuce lines (5.441 vs. 3.971 µg/100 mg fresh leaf mass for lettuce and tobacco, respectively). However tobacco is a model plant species to produce recombinant proteins, translation apparatus of lettuce has probably act better in case of GRFT expression.
The subsequent investigative steps involved comparative analysis of alternative GRFT-KDEL fusions (alone or with N-terminal Zera signal peptide) and their influence on transgene expression and protein build-up. Despite the high transcription rates noted in the transgenic line harboring GRFT-KDEL fusion without signal peptide in both species (TTK1 and TLK2), it did not lead to the detection of correspondingly pronounced GRFT levels. In contrast, fusing GRFT with the Zera signal peptide afforded a twofold increase in the recovery of the protein of interest, stably accumulated in protein body-like structures (Conley et al. 2011; Saberianfar et al. 2016). One of interesting characteristics of Zera fusion recombinant proteins is their potency to target and store in protein bodies. Higher accumulation of recombinant GRFT obtained with construct harboring N-terminal Zera signal peptide can probably be due to self-assembly and formation of GRFT inside the endoplasmic reticulum-derived protein bodies. However, immunocytochemistry and confocal microscopy is needed to confirm generation of protein bodies in lettuce and tobacco leaves.
Overall, the proportion of the herein retrieved codon-optimized and specifically targeted recombinant GRFT, relative to the total soluble protein content (89.42 µg/g FW−1 or 0.311% in the TLZ4 line) proved higher than that reported for other pharmaceutical proteins generated in stable nuclear-transformed transgenic lettuce: Hepatitis B Virus antigen, HBsAg, 40.11 µg/g FW−1 (Pniewski et al. 2017); G-binding protein A fused to a nucleotide sequence encoding pro-insulin, ProA-Pins, 0.13 TSP (Mohebodini et al. 2014); H1N1 influenza surface antigen, 0.045% TSP (Liu et al. 2012); fusion of the cholera toxin B subunit and the neutralizing epitope of the porcine epidemic diarrhea virus, sCTB-sCOE, 0.0065% TSP (Huy et al. 2011); recombinant plague antigen, F1-V, 0.08% TSP (Alvarez et al. 2010); cholera toxin B subunit, CTB, 0.24% TSP (Kim et al. 2006). Another issue that can be discussed is the comparison of nuclear-transformed lines with translastomics counterparts. Chloroplast transformation has been successfully used for high level production of important pharmaceutical. However, despite the numerous advantages of chloroplast transformation, all plant species are not amenable to the process, with the monocotyledons, including agronomically important grasses like rice or maize, proving especially problematic. On the other hand, cloning of plastid flanking sequences (such as accD and rbcL) and final species-specific chloroplast transformation sequences is a labor and time-consuming process (Vafaee et al. 2014).
Development of optimized recombinant expression systems is highly necessary for GRFT protein as only low level can be obtained from red algae Griffithsia sp. In our study, produced griffithsin in the nuclear-transformed lettuce and tobacco was detected in both monomeric form (13 kDa) and as a dimer (27 kDa). This was in agreement with the reports of O’Keefe et al. (2009) and Lotter-Stark et al. (2010) in which dimer GRFT was also produced by tobacco plant cells. Evidences are provided that the dimer form of GRFT is pivotal to the action of GRFT in HIV inhibition (Xue et al. 2013).
Recombinant production of GRFT has been mainly based on transient expression using virus-based vectors (Fuqua et al. 2015b; Hahn et al. 2015; Lotter-Stark et al. 2010; O’Keefe et al. 2009). Although, high level of recombinant GRFT has obtained in transient plant expression systems, presence of plant toxic metabolites and bacterial endotoxins as well as need for immediate process and complete purification of recombinant proteins have challenged these production platforms (Canto 2016). To overcome some of the aforementioned drawbacks, Fuqua et al. (2015b) could achieve more than 99% pure GRFT free of plant endotoxins and virus contaminant using a multistep methodology including heat (55 °C), magnesium chloride and bentonite treatments followed by a single chromatographic step. As the first report on stable expression of GRFT in a plant system, Vamvaka et al. (2016) could achieve 223 µg GRFT per gram dry rice seed mass. Although, we obtained lower GRFT level in lettuce and tobacco (up to 89.42 µg/g FW−1 in TLZ4 using Zera signal peptide), higher lettuce biomass (15–20 tons/ha leaf material) can probably led to obtaining higher purified recombinant protein in a similar platform.
For purification of recombinant GRFT from plants in an industrial scale, O’Keefe et al. (2009) and Fuqua et al. (2015b) used ceramic infiltration and Capto MMC chromatography, respectively. Similar to Vamvaka et al. (2016), we used immobilized metal affinity chromatography (IMAC) to purify recombinant GRFT but IMAC is an expensive purification method that is not economic for the industrial scale platforms. In this regard, as we could achieve higher GRFT accumulation with Zera signal peptide, further investigations can be proceed to establish a cost efficient manufacturing and an optimized purification method with focus on protein body fractionation and recombinant protein recovery.
GRFT can indirectly blocks the first step of HIV entrance to the CD4 cells by capturing the mannose-rich glycoproteins (gp120, gp41 and gp160) (Giomarelli et al. 2006). GRFT target a wide range of viruses, but due to binding to specific multiple regions on the gp120 glycoprotein, it particularly neutralize HIV strains (Mori et al. 2005). In this context, we tested gp120 binding ability of GRFT purified from one selected plant of T3 lettuce and tobacco generation and their binding potency was compared with GRFT produced in E. coli as control. The gp120 binding activity of GRFT was expectedly raised by increasing applied concentrations and GRFTE showed higher potency in lower levels (up to 1 ng). However gp120 binding ability of plant produced GRFT especially GRFTL was higher than GRFTE in the lower applied concentrations, they reached a stationary state at 1000 ng. In total, a similar binding potency similar to the bacterial protein was observed once the concentration of GRFT in the extracts was considered. Our results are in line with previous researches. For example, O’Keefe et al. (2009) as the first report on production of GRFT in a plant host showed that recombinant GRFT expressed in Nicotiana benthamiana had equal or even higher gp120 binding potency than GRFT produced in E. coli. On the other hand, crude rice endosperm extracts containing GRFT not only could bind to gp120 in a manner identical to that of purified GRFT without interfering endogenous plant lectins but also it neutralized HIV in whole cell and infectivity assays (Vamvaka et al. 2016). Later study suggested that crude plant extracts containing recombinant microbicides and lectins possess no cell toxicity and can even directly used for topical application without need for expensive purification methods.
Conclusion
GRFT has been introduced as a potent and efficient microbicide in the current clinical trials. This highlights need for its accessibility for at-risk countries. Therefore, development of optimized recombinant production systems for GRFT gives opportunity for further cost and labor analysis of transformation, expression, production and purification. Efficient molecular farming in leafy vegetables, affording high levels of transgene expression and protein accumulation, requires selection of appropriate host species and signal sequences, as well as application of strong promoters and optimized codon compositions based on plant preferences. In this study, we found that both lettuce and tobacco plants were optimal hosts for the generation of GRFT. However, compared to some other stable plant transformation reports mediated by agrobacterium, we could achieve higher amount of recombinant protein using Zera signal peptide, further investigation on protein body formation induced by Zera as well as complementary analyses including whole-cell HIV neutralization assay on lettuce- and tobacco derived GRFT can be performed.
Abbreviations
- BA:
-
Benzyladenine
- CaMV:
-
Cauliflower musaic virus
- CBA:
-
Carbohydrate binding agents
- DTT:
-
Dithiothreitol
- ELIZA:
-
Enzyme-linked immunosorbent assay
- GRFT:
-
Griffthsin
- HIV:
-
Human immunodeficiency virus
- IMAC:
-
Immobilized metal affinity chromatography
- MS:
-
Murashige and Skoog
- NAA:
-
Naphthalene acetic acid
- nptII:
-
Neomycin phosphotransferase
- PVDF:
-
Polyvinylidene difluoride
- SAR:
-
Scaffold-attachment regions
- TSP:
-
Total soluble protein
References
Allen GC, Spiker S, Thompson WF (2001) Use of matrix attachment regions (MARs) to minimize transgene silencing. Plant Mol Biol 43:361–376
Alvarez ML, Topal E, Martin F, Cardineau GA (2010) Higher accumulation of F1-V fusion recombinant protein in plants after induction of protein body formation. Plant Mol Biol 72:75–89
Appaiahgari MB, Kiran U, Ali A, Vrati S, Abdin MZ (2017) Plant-based edible vaccines: issues and advantages. In: Abdin MZ, Kiran U, Kamaluddin, Ali A (eds) Plant biotechnology: principles and applications. Springer, Singapore, pp 329–366
Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Buyel JF, Twyman RM, Fischer R (2017) Very-large-scale production of antibodies in plants: the biologization of manufacturing. Biotechnol Adv 35(4):458–465
Canto T (2016) Transient expression systems in plants: potentialities and constraints. In: Vega MC (ed) Advanced technologies for protein complex production and characterization. Springer, Cham, pp 287–301
Cao L, Diedrich JK, Kulp DW, Pauthner M, He L, Park S-KR, Sok D, Su CY, Delahunty CM, Menis S, Andrabi R, Guenaga J, Georgeson E, Kubitz M, Adachi Y, Burton DR, Schief WR, Yates JR III, Paulson JC (2017) Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat Commun 8:14954
Clemente T (2005) Nicotiana (Nicotiana tabacum, Nicotiana benthamiana). In: Wang K (ed) Agrobacterium protocols. Humana press, New York, pp 143–154
Conley AJ, Joensuu JJ, Richman A, Menassa R (2011) Protein body-inducing fusions for high-level production and purification of recombinant proteins in plants. Plant Biotechnol J 9:419–433
Curtis IS (2005) Lettuce (Lactuca satvia L.). In: Wang K (ed) Agrobacterium protocols. Humana press, New York, pp 449–458
Darqui FS, Radonic LM, López N, Hopp HE, López Bilbao M (2018) Simplified methodology for large scale isolation of homozygous transgenic lines of lettuce. Electron J Biotechnol 31:1–9
Dirisala VR, Nair RR, Srirama K, Reddy PN, Rao KRSS, Kumar NSS, Parvatam G (2016) Recombinant pharmaceutical protein production in plants: unraveling the therapeutic potential of molecular pharming. Acta Physiol Plant 39(1):18. https://doi.org/10.1007/s11738-016-2315-3
Dolgova AS, Dolgov SV, Nazipova NN, Maksimenko OG, Georgiev PG (2015) Arabidopsis termination elements increase transgene expression in tobacco plants. Plant Cell Tissue Organ Cult 120(3):1107–1116
Elvira-Matelot E, Martínez ÁE (2017) Diversity of RNA silencing pathways in plants. In: Dalmay T (ed) Plant gene silencing: mechanisms and applications. CABI Publication, Boston, pp 1–31
Fuqua JL, Hamorsky K, Khalsa G, Matoba N, Palmer KE (2015a) Bulk production of the antiviral lectin griffithsin. Plant Biotechnol J 13(8):1160–1168. https://doi.org/10.1111/pbi.12433
Fuqua JL, Wanga V, Palmer KE (2015b) Improving the large scale purification of the HIV microbicide, griffithsin. BMC Biotechnol 15:12
Gelvin SB (2017) Integration of agrobacterium T-DNA into the plant genome. Annu Rev Genet 51(1):195–217
Giomarelli B, Schumacher KM, Taylor TE, Sowder RC, Hartley JL, McMahon JB, Mori T (2006) Recombinant production of anti-HIV protein, griffithsin, by auto-induction in a fermentor culture. Protein Expr Purif 7:194–202
Habibi P, Prado GS, Pelegrini PB, Hefferon KL, Soccol CR, Grossi-de-Sa MF (2017) Optimization of inside and outside factors to improve recombinant protein yield in plant. Plant Cell Tissue Organ Cult 130(3):449–467. https://doi.org/10.1007/s11240-017-1240-5
Hahn S, Giritch A, Bartels D, Bortesi L, Gleba Y (2015) A novel and fully scalable Agrobacterium spray-based process for manufacturing cellulases and other cost-sensitive proteins in plants. Plant Biotechnol J 13(5):708–716
Heron SE, Elahi S (2017) HIV infection and compromised mucosal immunity: oral manifestations and systemic inflammation. Front Immunol 8:241
Huy NX, Yang MS, Kim TG (2011) Expression of a cholera toxin B subunit-neutralizing epitope of the porcine epidemic diarrhea virus fusion gene in transgenic lettuce (Lactuca sativa L.). Mol Biotechnol 48:201–209
Kim YS, Kim BG, Kim TG, Kang TG, Yang MS (2006) Expression of a cholera toxin B subunit in transgenic lettuce (Lactuca sativa L.) using Agrobacterium-mediated transformation system. Plant Cell Tissue Organ Cult 87:203–210
Liu CW, Chen JW, Kang CC, Wu CH, Yiu JC (2012) Transgenic lettuce (Lactuca sativa L.) expressing H1N1 influenza surface antigen (neuraminidase). Sci Hortic 139:8–13
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25(4):402–408
Llop-Tous I, Ortiz M, Torrent M, Ludevid MD (2011) The expression of a xylanase targeted to ER-protein bodies provides a simple strategy to produce active insoluble enzyme polymers in tobacco plants. PLoS ONE 6(4):e19474
Lotter-Stark T, Chakauya E, Kabamba A, Morris L, Rybicki E, Chikwamba R (2010) The effect of subcellular targeting on the expression and accumulation of Griffithsin in N. benthamiana. Paper presented at the 3rd Biennial Conference: Science Real and Relevant, Pretoria
Mitchell CA, Ramessar K, O’Keefe BR (2017) Antiviral lectins: selective inhibitors of viral entry. Antiviral Res 142:37–54
Mohebodini M, Jalali-Javaran M, Alizadeh H, Mahboudi F, Yarbakht M (2014) Agrobacterium-mediated transformation of lettuce (Lactuca sativa L.) to express IgG-binding protein A and human pro-insulin as a fusion protein. J Hortic Sci Biotechnol 89(6):719–725
Mori T, O’Keefe BR, Sowder RC, Bringans S, Gardella R, Berg S, Cochran P, Turpin JA, Buckheit RW, McMahon JB (2005) Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J Biol Chem 280:9345–9353
Mortimer CL, Dugdale B, Dale JL (2015) Updates in inducible transgene expression using viral vectors: from transient to stable expression. Curr Opin Biotechnol 32:85–92
Moustafa K, Makhzoum A, Trémouillaux-Guiller J (2016) Molecular farming on rescue of pharma industry for next generations. Crit Rev Biotechnol 36(5):840–850. https://doi.org/10.3109/07388551.2015.1049934
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue culture. 15:473–497
O’Keefe BR, Vojdani F, Buffa Y, Shattock RG, Montefiori DC, Bakke J, Mirsalis J, d’Andrea DA, Hume SD, Bratcher B, Saucedo CJ, McMahon JB, Pogue JP, Palmer KP (2009) Scaleable manufacture of HIV-1 entry inhibitor griffithsin and validation of its safety and efficacy as a topical microbicide component. Proc Natl Acad Sci USA 106:6099–6104
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acid Res 30:e36
Pniewski T, Czyż M, Wyrwa K, Bociąg P, Krajewski P, Kapusta J (2017) Micropropagation of transgenic lettuce containing HBsAg as a method of mass-scale production of standardised plant material for biofarming purposes. Plant Cell Rep 36(1):49–60
Rajeev Kumar S, Anunanthini P, Ramalingam S (2015) Epigenetic silencing in transgenic plants. Front Plant Sci 6:693
Saberianfar R, Sattarzadeh A, Joensuu JJ, Kohalmi SE, Menassa R (2016) Protein bodies in leaves exchange contents through the endoplasmic reticulum. Front Plant Sci 7:693. https://doi.org/10.3389/fpls.2016.00693
Schmidt S, Parott WA (2001) Quantitative detection of transgenes in soybean [Glycine max (L.) Merrill] and peanut (Arachis hypogaea L.) by real-time polymerase chain reaction. Plant Cell Rep 20:422–428
Singh R, Yadav R, Amla DV, Sanyal I (2016) Characterization and functional validation of two scaffold attachment regions (SARs) from Cicer arietinum (L.). Plant Cell Tissue Organ Cult 125(1):135–148
Tang W, Newton RJ, Weidner DA (2007) Genetic transformation and gene silencing mediated by multiple copies of a transgene in eastern white pine. J Exp Bot 58:545–554
Torrent M, Llop-Tous I, Ludevid MD (2009) Protein body induction: a new tool to produce and recover recombinant proteins in plants. Methods Mol Biol 483:193–208
Vafaee Y, Staniek A, Mancheno-Sonao M, Warzecha H (2014) A modular cloning toolbox for the generation of chloroplast transformation vectors. PLoS ONE 9(10):e110222
Vamvaka E, Arcalis E, Ramessar K, Evans A, O’Keefe BR, Shattock RJ, Medina V, Stoger E, Christou P, Capell T (2016) Rice endosperm is cost-effective for the production of recombinant griffithsin with potent activity against HIV. Plant Biotechnol J 14(6):1427–1437
Warzecha H (2008) Biopharmaceuticals from plants: a multitude of options for posttranslational modifications. Biotechnol Genet Eng Rev 25:315–330
World Health Organization (WHO) (2016) http://www.who.int/gho/hiv/epidemic_status/cases_all/en/
Xue J, Hoorelbeke B, Kagiampakis I, Demeler B, Balzarini J, LiWang PJ (2013) The griffithsin dimer is required for high-potency inhibition of HIV-1: evidence for manipulation of the structure of gp120 as part of the griffithsin dimer mechanism. Antimicrob Agents Chemother 57(8):3976–3989
Zhang Y, Liu Y, Zhang J, Wang G, Wang J, Liu Y (2015) Assessment of transgene copy number and zygosity of transgenic maize overexpressing Cry1Ie gene with SYBR® Green qRT-PCR. In Vitro Cell Dev Biol Plant 51(2):125–134
Zhao C-P, Guo X, Chen S-J, Li C-Z, Yang Y, Zhang J-H, Chen S-N, Jia Y-L, Wang T-Y (2017) Matrix attachment region combinations increase transgene expression in transfected Chinese hamster ovary cells. Sci Rep 7:42805
http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=4236. Accessed 14 Nov 2016
Acknowledgements
The authors are grateful to Dr. Heribert Warzecha and Dr. Agata Staniek for their constant encouragement for this work and valuable comments on the manuscript. This project was funded with funds from Vice Chancellor for research, University of Kurdistan (Research Grant Number 4/11351). We thanks Dr. Ali Fathi for his expert technical assistant for gp120 neutralization assay.
Author information
Authors and Affiliations
Contributions
HA conceived the work and designed experiments and contributed reagents. YA performed experiments, analyzed data and wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
There are no declare of interest statement.
Additional information
Communicated by Sergio J. Ochatt.
Rights and permissions
About this article
Cite this article
Vafaee, Y., Alizadeh, H. Heterologous production of recombinant anti-HIV microbicide griffithsin in transgenic lettuce and tobacco lines. Plant Cell Tiss Organ Cult 135, 85–97 (2018). https://doi.org/10.1007/s11240-018-1445-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-018-1445-2