Abstract
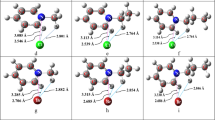
Density functional theory (DFT), MP2, and couple cluster ab initio methods were employed to investigate the microsolvation of 3-methyl-4-pyrimidone (3M4P) surrounded by methanol (MeOH) molecules. Structures are analyzed based on hydrogen bonds with a focus on relative energies, interaction energies, hydrogen bond cooperativity, hydrogen bonding geometries, and redshifts in the frequencies of O–H and C=O stretching modes. Our results show that there is no preferential orientation of MeOH attacks on the carbonyle site of 3M4P; both trans and cis 3M4P-MeOH complexes have same chance to be observed. cis 3M4P-MeOH and 3M4P-MeOH complex in which MeOH is located on N lie 0.56 and 3.11 kJ/mol at CCSD(T)/6-31+G(d,p) (0.63 and 1.67 kJ/mol at MP2/6-311++G(d,p)) above trans 3M4P-MeOH. MeOH dimers form more stable 3M4P-(MeOH)2 complexes compare to 3M4P-(MeOH)2 complexes in which individual MeOH molecules bind to carbonyl and N. Relative energies of 3M4P-(MeOH)3 computed using various DFT methods point out the complex formed by linear MeOH trimer along methyl group of 3M4P (cis 3M4P-(MeOH)3) as lowest. Carbonyl group is predicted as preferential site for hydrogen bond interaction. Besides O–H…O and O–H…N hydrogen bonds, 3M4P-(MeOH)2 and 3M4P-(MeOH)3 complexes are also stabilized by H–O…H–C weak interactions.




Similar content being viewed by others
References
Alberts B, Johnson A, Lewis J, et al. (2002) Molecular biology of the cell, 4th edn. Garland Science, New York,
Lodish H, Berk A, Zipursky SL, et al. (2000) Molecular cell biology, 4th edn. W. H. Freeman, New York,
Jeffrey GA, Saenger W (1991) Hydrogen bonding in biological structures. Springer-Verlag, Berlin,
Jeffrey GA (1997) An introduction to hydrogen bonding. Oxford University Press, New York,
Grabowski SJ (ed.) (2006) Hydrogen bonding—new insights. Springer, New York,
Scheiner S (1997) Hydrogen bonding, a theoretical perspective. Oxford University Press, New York,
Szatyłowicz H, Sadlej-Sosnowska N (2010) J Chem Inf Model 50:2151–2161
Matta CF, Castillo N, Boyd RJ (2006) Extended weak bonding interactions in DNA: π-stacking (base-base), base-backbone, and backbone-backbone interactions J Phys Chem B 110:563–578
Kasende O, Zeegers-Huyskens T (1981) J Mol Struct 75:201–207
Kasende O, Zeegers-Huyskens T (1984) J Phys Chem 88:2636–2642
Kasende O, Zeegers-Huyskens T (1984) Spectrosc Lett 17:783–801
Muzomwe M, Boeckx B, Maes G, Kasende OE (2011) S Afr J Chem 64:23–33
Muzomwe M, Maes G, Kasende OE (2012) Nat Sci 4:286–297
Kasende OE, Muya JT, Broeckaert L, Maes G, Geerlings P (2011) J Phys Chem A 116:8608–8614
Szalewicz K (2012) Wiley interdisciplinary reviews: computational molecular science, vol 2, pp. 254–272
Shao Y, Gan Z, Epifanovsky E, Gilbert ATB, et al. (2015) Mol Phys 113:184–215
Weinhold F, Landis CR (2012) Discovering chemistry with natural bond orbitals. John Wiley & Sons, Hoboken, New Jersey,
Gaussian 09, Revision D.01, M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery, Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski, and D. J. Fox, Gaussian, Inc., Wallingford CT, (2009).
GaussView, Version 5, Dennington, R.; Keith, T.; Millam, J., Semichem Inc., Shawnee Mission, KS (2009).
Geerlings P, Proft FD, Langenaeker W (2003) Conceptual density functional theory Chem Rev
Murray JS, Politzer P (2011) The electrostatic potential : an overview Commput Mol Sci 1:153–163
Acknowledgements
JT Muya thanks the Korea Research Fellowship program for the financial support and Prof. Carol Parish for the support and permissions to use the facilities and computing resources of the University of Richmond.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary materials
ESM 1
Table S1. Selected bonds and angles formed by MeOH in 3M4P-MeOH (1b–d) computed using various DFT and MP2 at 6-311++G(d,p) and CCSD/6-31G+(d,p). The B3LYP and CCSD values given in parenthesis were obtained with 6-31+G(d) and 6-31G(d) basis sets, respectively. Table S2. SAPT2 interaction energies in kJ/mol components computed at HF/6-31+G(d,p) of 3M4P-MeOH. Table S3. Red shifted of O–H and C=O vibrational frequencies (Δν in cm−1) involved in the hydrogen bond interaction between 3M4P and MeOH in 3M4P-(MeOH)1–3 complexes calculated at B3LYP/6-31++G(d,p). In parenthesis are red shifted of O–H vibrator between MeOH molecules. Fig. S1. Labels of atoms in 3M4P-MeOH (a) and 3M4P-(MeOH)2 (b) complexes used for E(2) NBO analysis in Table 4. (DOCX 149 kb)
Rights and permissions
About this article
Cite this article
Muzomwe, M., Muya, J.T., Hoeil, C. et al. Theoretical study on coordination of methanol clusters to 3-methyl-4-pyrimidone. Struct Chem 29, 1–8 (2018). https://doi.org/10.1007/s11224-017-0992-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-017-0992-1