Abstract
Weeds are a significant threat to agricultural productivity and the environment. The increasing demand for sustainable weed control practices has driven innovative developments in alternative weed control technologies aimed at reducing the reliance on herbicides. The barrier to adoption of these technologies for selective in-crop use is availability of suitably effective weed recognition. With the great success of deep learning in various vision tasks, many promising image-based weed detection algorithms have been developed. This paper reviews recent developments of deep learning techniques in the field of image-based weed detection. The review begins with an introduction to the fundamentals of deep learning related to weed detection. Next, recent advancements in deep weed detection are reviewed with the discussion of the research materials including public weed datasets. Finally, the challenges of developing practically deployable weed detection methods are summarized, together with the discussions of the opportunities for future research. We hope that this review will provide a timely survey of the field and attract more researchers to address this inter-disciplinary research problem.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Due to their competition with crops for water, nutrients and sunlight, weeds are a significant threat to agricultural productivity (Gharde et al., 2018; Llewellyn et al., 2016). Weed control in conservation cropping systems is reliant on the use of herbicides due to the lack of suitable alternative weed control options that do not interfere with the principles of minimum tillage and residue retention and thus the substantial benefits of this system. The site-specific approach to weed control (SSWC) creates the opportunity to alleviate this threat through the precision application of alternative weed control treatments such as lasers, electrical weeding, waterjet cutting etc. (Coleman et al., 2019). However, to achieve selective in-crop weed control, that avoids crop damage with these alternative treatments accurate weed recognition in all conditions is essential.
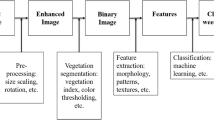
The complex and highly variable cropping environment is a significant barrier to the development of robust weed recognition algorithms (Olsen et al., 2019). Plant morphologies as influenced by genetics and the environment vary considerably both between plant species (crop and weed) as well as within these species to create a substantial challenge to the development of weed recognition algorithms. It is possible that extent of morphological variability and changing complexity will require differing weed recognition approaches based for example on species (crop and weed), growth stage, environment and combinations of these influences on plant growth. In scenarios where there are substantial differences in plant morphology the weed recognition challenge will be simpler than those where the crop and weed plants are very similar. The majority of previous weed recognition methods were based on conventional computer vision and machine learning techniques in both colour and multispectral imagery. These approaches often follow a pipeline in which hand-crafted image features play a primary role (e.g., shape features (Charters et al., 2014)). As a result, developing such pipelines is labour intensive and images are required to be captured within well-defined conditions (Fig. 1).
Fortunately, due to the great success of deep learning in many vision tasks, hand-crafted features are no longer required to derive promising results. Instead, deep representations of an input image can be obtained using deep learning, which are relevant to the task at hand. For weed recognition, four types of deep learning approaches are used as illustrated in Fig. 2: (a) image classification identifies the weed or the crop species contained in an image; (b) object detection identifies the per-weed locations of the plants within an image; (c) semantic segmentation conducts pixel-wise classification of individual weed classes and (d) instance segmentation further identifies the instance each pixel belongs to. As most of the deep learning-based weed recognition studies are based on existing and well-known deep architectures, those relevant to weed recognition, including their building blocks and contributions, are introduced first. Next, more than 30 deep learning-based weed recognition studies are discussed in terms of their architectures, goals and performance. In addition, as deep learning based weed recognition research often requires a large volume of annotated data, we provide the details of current publicly available weed datasets and benchmarking metrics.
Expanding on the research of existing well-known architectures, we present other fine-grained and alternative architectures that may offer advantages in terms of the recognition performance for future research. In practice, there are still limitations and challenges for current weed recognition research to provide weed control in large-scale crop production systems. Therefore, deep learning mechanisms which could further improve the efficiency and effectiveness of weed control including real-time inference, weakly-supervised learning, explainable learning and incremental learning techniques are discussed.
In summary, this review aims to: (1) investigate deep learning techniques related to weed control; (2) summarize current deep learning-based weed recognition research including architectures, research materials and evaluation methods; (3) identify further challenges and improvements for future research with deep learning-based solutions.
The remainder of the review is organized as follows. In “Overview of Deep Learning Techniques” section, deep learning architectures related to weed control are introduced. “Deep Learning for Weed Recognition” section provides a discussion of the existing deep learning based weed detection studies. In addition, public datasets and evaluation metrics for benchmarking are summarised. “Discussion” section considers the challenges for weed detection and potential deep learning solutions. Finally, “Conclusion” section summarises this review.
Overview of deep learning techniques
In this section, the theory of deep learning techniques is introduced including the deep learning building blocks and architectures relevant for weed detection.
Machine learning
Machine learning (ML) algorithms are a class of algorithms that ’learn’ to perform a specific task given sample data (i.e., training data). These algorithms are not explicitly programmed with rules or instructions to fulfil the task. In general, a set of samples for a ML task \({\textbf{D}}=\{({\textbf{x}}_{i}, {\textbf{y}}_{i})\}\) can be obtained, where \({\textbf{x}}_{i} \in {\textbf{R}}^{p}\) is the observed features describing the characteristics of the i-th sample and \({\textbf{y}}_{i}\) is the associated output. In general, for \(({\textbf{x}}, {\textbf{y}})\in {\textbf{D}}\), it can be costly and time-consuming to obtain \({\textbf{y}}\), whilst \({\textbf{x}}\) is convenient to collect. Therefore, it is expected to learn a model \(f_{\mathbf {\Theta }}({\textbf{x}})\) that maps input values to the target variable \(\hat{{\textbf{y}}}\) as close as possible, where \(\mathbf {\Theta }\) is a set containing parameters of the model. Optimization methods can be used to find the best set of model parameters, \(\hat{\mathbf {\Theta }}\), to minimize the difference between the predicted output \(\hat{{\textbf{y}}}=f_{\hat{\mathbf {\Theta }}}({\textbf{x}})\) and the ground truth \({\textbf{y}}\). In regard to the form of \({\textbf{y}}\), machine learning problems can be differentiated as classification problems if the value of \({\textbf{y}}\) is categorical, or regression problems if the value of \({\textbf{y}}\) is continuous.
In the past decades, various machine learning models have been proposed (e.g. support vector machines (Cortes & Vapnik, 1995)). However, these methods require carefully devised hand-crafted features. Thanks to the recent growth in computational capacity and the availability of a large amount of training data, deep learning automatically integrates feature extraction and modelling together, and promising performance gains have been observed in many varied tasks. Figure 1 provides an illustration of the difference between conventional machine learning vs. deep learning. In the following subsections, the details of deep learning are introduced.
Neural networks
The model \(f_{\mathbf {\Theta }}\) in machine learning can be chosen as a neural network (NN) (Schmidhuber, 2015), which contains an interconnected group of nodes (i.e., artificial neurons) inspired and simplified by the biological neural networks in animal brains. The most well-known neural network architectures are the multi-layered perceptrons (MLPs), shown in Fig. 3a. This architecture organizes nodes into groups of layers and connects nodes between neighbouring layers. In detail, the computations of the k-th layer can be written as:
where \({\textbf{x}}^{(k)}\in {\textbf{R}}^{p^{(k)}}\) is the input of the k-th layer which can be viewed as \(p^{(k)}\) nodes of the neural network; \({\textbf{W}}^{(k)}\in {\textbf{R}}^{p^{(k)}\times p^{(k-1)}}\) with the bias vector \({\textbf{b}}^{(k)}\in {\textbf{R}}^{p^{(k)}}\) represents a linear transform of the input signal which introduces full connectivity between the \((k-1)\)-th layer and the k-th layer; \(\sigma\) is an activation function which introduces a non-linearity to the output, allowing complex representations. In particular, \({\textbf{x}}^{(0)}\) is the input feature of a sample in D.
The layer defined in Eq. (1) can also be referred to as a fully connected (FC) layer. By stacking multiple FC layers, neural networks are able to formulate more complex representations of the input. To obtain predictions, computations are conducted from the first (input) layer to the last (output) layer, which is known as a forward propagation stage. To optimize the parameters \(\mathbf {\Theta }=\{{\textbf{W}}^{(k)}, {\textbf{b}}^{(k)}\}\) of a neural network, a backward propagation stage updates the parameters in an inverse order. Recently, more mechanisms and architectures have been proposed for constructing deeper neural networks, of which the ones related to weed recognition are reviewed in the rest of this section.
Convolution neural networks
Inspired by the biological processes of animal visual cortex, convolution neural networks (CNNs) reduce the challenges of training deep models for visual tasks (Gu et al., 2018). Convolution layers are the key components of CNNs, as illustrated in Fig. 3b. It involve partial connections compared with fully connected layers of MLPs, where each node focuses on a local region of the input. In detail, denote \(W^{(k)} = \{W_{1}^{(k)}, W_{2}^{(k)},\ldots , W_{C^{(k)}}^{(k)}\}\) to represent a series of \(C^{(k)}\) convolution filters and the computations of the k-th convolution layer can be written as:
where \(*\) represents a convolution operator and \(\sigma\) is an activation function; \({\textbf{x}}^{(k-1)}\) is the input feature map containing \(C^{(k-1)}\) channels and the output feature map is \({\textbf{x}}^{(k)} = ({\textbf{x}}_{1}^{(k)},\ldots ,{\textbf{x}}_{C^{(k)}}^{(k)})\) containing \(C^{(k)}\) channels. A convolution layer can be viewed as a special case of FC layers with a sparse weight matrix.
Convolution layers often reduce the input spatial size but increase the number of channels. For some applications, recovery from a deep representation to the original input size is required. For this purpose, deconvolution (transpose convolution) operations were used. Readers can refer to (Zeiler et al., 2010) for more details.
Graph neural networks
Whereas most neural networks were designed for processing vectorized data, there are a wide range of applications involving non-vectorized data. Graph neural networks (GNNs) were devised for graph inputs. One commonly used GNN is graph convolution neural networks (GCNNs), which is a generalisation of the conventional CNNs by involving adjacency patterns (Bruna et al., 2013). In detail, a particular form to compute graph convolutions in k-th layer can be written as:
where \({\textbf{X}}^{(k)}\in {\textbf{R}}^{n\times p^{(k)}}\) represents the vertex features of n vertices in a graph, \({\textbf{A}}\in {\textbf{R}}^{n\times n}\) is an adjacency matrix to illustrate the relations between vertices, \({\textbf{W}}^{(k)}\in {\textbf{R}}^{p^{(k-1)}\times p^{(k)}}\) contains trainable weights and \({\textbf{b}}^{(k)}\) is a bias vector. Instead of using pre-defined adjacency matrix, graph attention network (GAT) estimated edge weights of the adjacency in line with the vertex features ( Veličković et al., 2018). Recently, various methods were proposed to focus on some graph characteristics, which could not be captured by GCNNs (e.g. longest circle in (Garg et al., 2020)).
Deep learning architectures
Following the above discussed neural networks for deep learning, various deep learning architectures can be constructed in line with different target applications. In terms of weed detection tasks, four categories of deep neural network architectures are summarized, including image classification, object detection, semantic segmentation, and instance segmentation.
Image classification
Image classification tasks focus on predicting the category (e.g. weed species) of the object in an input image. Input images can be viewed as spatially organized data, and many CNN based architectures have been proposed for classifying them into a specific class or category. AlexNet, which consists of 5 convolution layers and 3 fully connected layers was first adopted for large scale image classification (Krizhevsky et al., 2012). Convolution layers (potentially with other mechanisms) were used to formulate deep representations from input images and FC layers were further used to generate output vectors in line with the categories involved. VGG (Simonyan & Zisserman , 2014) further introduced convolution filters with a \(3 \times 3\) perception field to substitute each convolution filter with a large perception field to learn a deeper representation and reduce the computational costs. InceptionNet was further proposed to introduce filters of multiple sizes at the same level to characterize the salient regions in an image, which can have extremely large variations in size (Szegedy et al., 2017). With the growing depth of the CNN architectures, short-cuts between the layers alleviate the gradient vanishing issues (Huang et al., 2017), such as ResNet and DenseNet. NASNet (Zoph et al., 2018) was obtained by architecture search algorithms.
Object detection
Object detection aims to identify the positions and the classes of the objects contained in an image. Generally, various CNN architectures for image classification can be used as backbones to learn deep representations and specific output layers can be introduced to obtain object-level annotations including positions and categories. For example, R-CNN and its improvements such as Faster R-CNN (Ren et al., 2015) were proposed with a two-stage scheme where the first stage generates regional proposals and the second stage predicts the positions and labels for those proposals. One-stage methods were also explored to perform object detection with less latency. For example, YOLO (Redmon et al., 2016) (You Only Look Once) was proposed by treating object detection as a regression problem, of which the output is a feature map containing the positions and labels of each pre-defined grid cell. Single shot multi-box detector (SSD) (Liu et al., 2016) introduced feature maps of multiple scales and prior anchor boxes of different ratios. As class imbalance is one of the critical challenges for one-stage object detection, RetinaNet with focal loss was proposed (Lin et al., 2017). Note that pre-defined anchor boxes played an important role for most of the above mentioned methods. To avoid the significant computational costs to compute such anchor boxes, anchor-free methods were also investigated in (Tian et al., 2019).
Semantic segmentation
Semantic segmentation focuses on the pixel-wise (dense) predictions of an image, by which the category of each pixel is identified. In general, semantic segmentation uses fully convolution networks (FCNs), which were first explored in (Long et al., 2015). These studies often involved an encoder-decoder scheme: the encoder formulates a latent representation of an image through convolutions and the decoder focuses on upsampling of the latent representation to the original image size for dense predictions. By increasing the capacity of the decoder, U-Net (Ronneberger et al., 2015) was proposed with promising performance for medical images. SegNet (Badrinarayanan et al., 2017) additionally involved pooling indices for its decoder, compared with the general encoder-decoder models to use the pooled values only, to perform non-linear upsampling to keep the boundary information. Pyramid scene parsing network (PSPNet) and different-region-based context aggregation by a pyramid pooling module (Zhao et al., 2017) exploited the capability of global context as a superior framework for pixel-level predictions. Instead of following a conventional encoder-decoder scheme, DeepLab models adopted atrous convolutions to reduce the downsampling operations with a large reception field (Chen et al., 2018).
Instance segmentation
Instance segmentation aims to output both the class and class instance information for individual pixels. Instance segmentation methods were initially devised in a two-stage manner by focusing on two separated tasks: object detection and semantic segmentation. For example, Mask R-CNN (He et al., 2017) was proposed using a top-down design, which first conducts object detection task to locate the bounding boxes of each instance and next within each bounding box undertakes semantic segmentation. Bottom-up methods were also investigated, which first conduct semantic segmentation and use clustering or metric learning to obtain different instances (e.g. (Papandreou et al., 2018)). Two-stage methods require accurate results from each stage and the computation cost of two-stage methods could be expensive. Therefore, single shot methods were explored. Anchor-based methods YOLACT (Bolya et al., 2019) introduced two parallelized tasks to an existing one-stage object detection model including a dictionary of non-local prototype masks over the entire image and predicting a set of linear combination coefficients per instance. An anchor-free method fully convolutional instance-aware semantic segmentation (FCIS) was proposed based on FCNs by introducing position-sensitive inside/outside score maps (Li et al., 2017). PolarMask (Xie et al., 2020a) conducted instance center classification and dense distance regression in a polar coordinate, which is a much simpler and flexible framework. Very recently, BlendMask (Chen et al., 2020) inspired by FCIS introduced a blender module to effectively combine instance-level information and semantic information with low-level fine-granularity.
Deep learning for weed recognition
In this section, deep learning based weed recognition studies are summarized covering four 4 approaches: image classification, object detection, semantic segmentation and instance segmentation. Before reviewing those approaches, research data, data augmentation and evaluation metrics used in these studies are reviewed first to provide a context in the field.
Weed data
Weed data is the foundation for developing and benchmarking weed recognition methods, and sensing technologies will determine what weed data can be acquired and what weed management practice can be developed (Machleb et al., 2020). While various sensing techniques like ultrasound, light detection and ranging (LiDAR) and optoelectronic sensors were used for simple differentiation between weeds and crops, image-based weed recognition has gained increasing interests due to the advances of various imaging techniques.
Multispectral imaging captures light energy within specific wavelength ranges or bands of the electromagnetic spectrum, which can capture information beyond visible wavelength (Farooq et al., 2018). For example, hyperspectral images consist of many contiguous and narrow bands; near infrared (NIR) imaging uses a subset of the infrared band, as the pigment in plant leaves, chlorophyll, strongly absorbs red and blue visible light and reflects near infrared.
Driven by the low cost RGB cameras and the significant progresses in computer vision, RGB images have been increasingly used (e.g. (Olsen et al., 2019)). In addition, some studies involved the fusion of depth (the distance between the image plane and each pixel) and RGB images using sensors such as the Kinect v2. The result improved segmentation from 76.4% for colour-only, to 96.6% for broccoli (Gai et al., 2020).
The availability of rich public datasets in the field plays a key role in facilitating the development of new algorithms specific to weed recognition tasks. In recent years, a number of in-crop weed datasets have been made public, as shown in Fig. 4 and will be reviewed in the rest of this section.
Sample weed images from several public datasets. a Carrot-Weed (Lameski et al., 2017), b CWF-788 (Li et al., 2019), c CWF-ID (Haug & Ostermann, 2014), d DeepWeeds (Olsen et al., 2019), e GrassClover (Skovsen et al., 2019), f Plant Seedlings Dataset (Giselsson et al., 2017), g Sugar Beets 2016 (Chebrolu et al., 2017), h Sugar Beet/Weed Dataset (Sa et al., 2017), i Weed-Corn/Lettuce/Radish (Jiang et al., 2020)
-
Bccr-segset (Le Nguyen Thanh et al., 2019) contains 30 000 RGB images with pixel-wise annotations collected of canola (Brassica napus), maize (Zea mays) and wild radish (Raphanus raphanistrum). The images were acquired using an gantry-system mounted above an indoor growth facility across multiple growth stages.
-
Carrot-Weed (Lameski et al., 2017) contains 39 RGB images collected with a 10 MP (Mega Pixel) phone camera under variable light conditions of young carrot (Daucus carota subsp. sativus) seedlings in the Republic of Macedonia. Pixel-level annotations were provided of three categories: carrots, unspecified weeds and soil (https://github.com/lameski/rgbweeddetection).
-
Crop/Weed Field Image Dataset (CWFID) (Haug & Ostermann , 2014) comprises 60 top-down field images of carrots with intra-row and close-to-crop weeds captured by RGB cameras. Pixel-level annotations are provided for crop versus weed discrimination of 162 carrot plants and 332 weeds in total (https://github.com/cwfid).
-
CWF-788 (Li et al., 2019) is a field image dataset containing 788 RGB images collected from cauliflower (Brassica oleracea var. botrytis) fields with high weed pressure. It was collected for semantic segmentation of the cauliflower plants from the background (combining both weeds and soil) with manually segmented annotations (https://github.com/ZhangXG001/Real-Time-Crop-Recognition).
-
DeepWeeds (Olsen et al., 2019) was collected from remote rangelands in northern Australia for weed specific image classification. It includes 17 509 images of 8 types of target weed species with various off-target plants native to Australia. The target weed species include chinee apple (Ziziphus mauritiana), lantana (Lantana camara), parkinsonia (Parkinsonia aculeata), parthenium (Parthenium hysterophorus), prickly acacia (Vachellia nilotica), rubber vine (Cryptostegia grandiflora), siam weed (Chromolaena odorata) and snake weed (Stachytarpheta spp.). For each target weed species (positive class), around 1000 images were obtained; off-target flora and backgrounds not containing the weeds of interest are collected as a single negative class of 9106 images. It was collected from eight different locations with an attempt to balance scene bias images of the target species and negative cases were collected at each location in similar quantities (https://github.com/AlexOlsen/DeepWeeds).
-
Grass-Broadleaf (Dyrmann et al., 2016a) contains 22 different plant species at early growth stages, which was constructed by combining 6 image datasets. In total, 10 413 RGB images were included. Note that image background was removed in this dataset and each image only contains one individual plant.
-
GrassClover (Skovsen et al., 2019) is a diverse image and biomass dataset, of which 8 000 synthetic RGB images are provided with pixel-wise annotations for semantic segmentation based weed recognition studies. The dataset was collected in an outdoor field setting including 6 classes: unspecified grass species, white clover (Trifolium repens), red clover (Trifolium pratense), shepherd’s purse (Capsella bursa-pastoris), unspecified thistle, dandelion (Taraxacum spp.) and soil. In addition, 31 600 unlabelled images were provided for pre-training, weakly-supervised learning and unsupervised learning (https://vision.eng.au.dk/grass-clover-dataset).
-
Plant Seedling Dataset (Giselsson et al., 2017) contains 960 unique plants at several growth stages in RGB images for species including blackgrass (Alopecurus myosuroides), charlock (Sinapis arvensis), cleavers (Galium aparine), common chickweed (Stellaria media), wheat, fat hen (Chenopodium album), loose silky-bent (Apera spica-venti), maize (Zea mays), scentless mayweed, shepherd’s purse, small-flowered cranesbill (Geranium pusillum) and sugar beet (Beta vulgaris var. altissima) (https://vision.eng.au.dk/plant-seedlings-dataset).
-
Soybean/Grass/Broadleaf/Soil (dos Santos et al., 2017) comprises 15 336 segments of soybean (Glycine max), unspecified grass weeds, unspecified broadleaf weeds and soil. The segments were extracted using the simple linear iterative clustering (SLIC) superpixel algorithm on 400 images collected with an unmanned aerial vehicle (UAV)-mounted RGB camera.
-
Sugar Beets 2016 (Chebrolu et al., 2017) was collected from agricultural fields with pixel-wise annotations for three classes: sugar beet, weeds, and soil. This dataset contains 1600 images, of which 700 images were captured at first and 900 images were captured after a four-week period. Both RGB-D and multispectral images were provided, which is helpful to explore the effectiveness of different modalities for weed recognition and to construct multi-modal learning methods (http://www.ipb.uni-bonn.de/data/sugarbeets2016).
-
Sugar Beet/Weed Dataset (Sa et al., 2017) contains 155 multispectral images (near-infared 790 nm, red 660 nm) plus normalised difference vegetation index (NDVI) with pixel-wise labelling for sugar beet, weeds and soil from a controlled field experiment (https://github.com/inkyusa/weedNet).
-
Weed-AI is an open-source weed dataset upload and download platform that standardises metadata reporting with the WeedCOCO annotation format and centralises weed datasets. An annotation platform built on the computer vision annotation tool (CVAT) has been integrated. Weed-AI currently contains 17 datasets (including DeepWeeds) with 20 891 images. (https://weed-ai.sydney.edu.au/about).
-
Weed-Corn/Lettuce/Radish (Jiang et al., 2020) contains 7200 RGB images with image-level annotations. It includes three subsets: the maize dataset (1,200 images) was collected from a corn field with four different weed species (4800 images) including Canada thistle (Cirsium arvense), fat hen, bluegrass (Poa spp.) and sedge (Carex spp.); the lettuce dataset was collected from a vegetable field of two plant classes including lettuce (500 images) and weeds (300 images); the radish dataset contains 200 radish images and 200 weeds images (Lameski et al., 2017) (https://github.com/zhangchuanyin/weed-datasets).
Whilst these datasets provide useful imagery and annotation data for benchmarking, there is a lack of consistency and details in metadata reporting standards and contextual information. An understanding of weed species, beyond a simple awareness of the difference to crops, is important in creating opportunities to deliver specific weed control treatments. For example, contextual understanding of crop growth stages, presence/absence of stubble will assist in developing algorithms capable of handling variability across different conditions.
Data augmentation
Due to the laborious nature of developing annotated datasets within weed control contexts, existing datasets are often not large enough and do not reflect sufficient diversity in conditions. A significant risk for deep learning using small datasets is overfitting, where the model performs well on the training set but performs poorly when being deployed in the fields. To address this issue, various data augmentation strategies were adopted to enlarge the size and the quality of the training sets such as random cropping, rotation, flipping, color space transformation, noise injection, image mixing, random erasing and generative approaches. Readers can refer to (Shorten & Khoshgoftaar, 2019) for more details.
Evaluation metrics
A number of metrics have been utilised to evaluate the desktop performance of weed recognition algorithms. The definition of these metrics may differ in terms of different types of recognition approaches. The focus on desktop-based evaluation metrics instead of real-world field evaluation metrics is seen as a short coming of current methods (Salazar-Gomez et al., 2021). Nevertheless, the metrics are the current standard for comparison.
For binary image classification whereby the classification result of an input sample is labelled either as a positive (P) or a negative (N) case, 4 possible outcomes can be derived: (1) If a positive sample is classified as positive, the prediction is correct and defined as true positive (TP). (2) If a negative sample is classified as positive, the prediction is false positive (FP). (3) If a negative sample is classified as negative, the prediction is true negative (TN). (4) If a positive sample is classified as negative, the prediction is false negative (FN).
Based on these definitions, some widely used evaluation metrics can be defined for benchmarking the performance of different algorithms. Accuracy measures the proportion of the correct predictions (#TP + #TN) over all the predictions (#P + #N). Sensitivity, also known as recall, measures the proportion of the correctly predicted positive cases (#TP) over all the positive cases (#TP + #FN). It indicates the likelihood that the algorithm identifies all weeds. A low sensitivity would suggest that a large number of weeds are missed, while a sensitivity rate 1 indicates that all weeds are successfully detected. Precision measures the proportion of the correctly predicted positive cases (#TP) over all predicted positive cases (#TP + #FP). For weed detection, a high precision indicates low off-target or crop damage. Specificity measures the proportion of the correctly predicted negative cases (#TN) over all predicted negative cases (#TN + #FP). A low specificity suggests that an algorithm is delivering an control option towards crops. F-score (also known as F\(_{1}\)score) combines the precision and the recall values by treating them with equal importance:
As a binary classification model generally outputs continuous predictions, a threshold is required to judge the predicted labels: if the score is beyond the threshold, the corresponding sample is predicted as a positive case; otherwise the sample is predicted as a negative case. By varying the threshold, trade-offs among some metrics can be made. A receiver operating characteristic curve (ROC curve) illustrates the diagnostic ability of a binary classification model by plotting the sensitivity against the 1-specificity at various threshold settings. A precision-recall curve (PR curve) shows plots the precision against the recall. A large area under these curves (AUC) often indicates a model of high quality. For multi-class classification, most of the these metrics can be computed class by class and the mean of these metrics can be used.
For an object detection task with only one class, a sample is associated with an object in a bounding box. For a predicted bounding box, intersection over union (IoU) is defined as the area of the intersection divided by the area of the union of the predicted bounding box and a ground truth bounding box. Given a threshold \(\theta\), if the confidence value of a predicted bounding box is beyond \(\theta\) and the IoU against the ground truth bounding box is beyond 0.5, the predicted bounding box is regarded as a TP case; if the confidence is beyond \(\theta\) and the IoU is less than 0.5, it is regarded as a FP case; if the confidence is less than \(\theta\) and the IoU is less than 0.5, it is regarded as a TN case; if the confidence is less than \(\theta\) and the IoU is beyond 0.5, it is regarded as a FN case. Next, the precision and recall values can be defined to measure the quality of detection results. By varying \(\theta\), a PR curve can be obtained and average precision (AP): \(\int _{0}^{1} p(r)\textrm{d}r\) is used to summarize the quality of the PR curve, where p(r) indicates the precision value p corresponding to the recall values r for a particular IoU threshold. In practice, different estimations for AP are adopted such as the AUC of the PR curve. Different IoU threshold values other than 0.5 can also be used to select the bounding boxes from the candidates and the corresponding AP can be obtained. For example, AP\(_{50}\) and AP\(_{75}\) define the AP with IoU threshold 0.5 and 0.75, respectively. For multi-class object detection problems, these metrics can be computed for each class individually and a mean average precision (mAP) can be obtained over all classes.
In segmentation tasks, a sample can be viewed as a pixel. The metrics such as (mean) accuracy (mAcc), recall, precision and F-score discussed above can be derived in a similar manner. By organizing the pixels belonging to the same class as regions, the concepts such as mAP and mIoU can be derived as well.
Weed recognition methods
Existing studies on weed recognition can be organized into four categories in terms of the approaches they used: weed image classification, weed object detection, weed object segmentation or weed instance segmentation. Each approach represents a trade-off between algorithm complexity (i.e., speed of inference, training data difficulty) and the level of in-field recognition granularity that is provided as an output. We suggest that the selection of an approach will depend on the crop-weed combination, the weed control treatment scenario and the training and on-vehicle inference constraints. Table 1 provides an overview of these approaches from multiple perspectives such as computational cost and speed measured by floating-point operations per second (FLOPS), model size, power consumption, annotation intensity, recognition granularity and potential treatments. Tables 2, 3, 4 summarise the major studies of the first three categories, whilst instance segmentation based weed recognition has emerged recently.
Weed image classification
This approach aims to achieve image-level weed recognition, determining of what species or crop/non-crop plants an image contains. An early deep learning based study (Dyrmann et al., 2016a) devised a residual CNN for multi-class classification. On their proposed Grass-Broadleaf dataset which contains 10 413 RGB crop-weed images of 22 weed species, an accuracy 86.2% was achieved. A variant principal component analysis (PCA) network was proposed for classifying 91 classes of weed seeds using RGB images, and an accuracy 90.96% was achieved. AlexNet was adopted to classify RGB images from the public dataset - Grass-Broadleaf (dos Santos et al., 2017), which achieved an accuracy 99.5%. Although these results look promising, the plants or seeds were well segmented and the field or natural background information was limited, which could lead to failures under real field conditions.
More recently, a hybrid model of AlexNet and VGGNet was proposed. It was evaluated on a public plant seedling dataset containing RGB images of 3 crop species and 9 weed species at an early growth stage and achieved an accuracy 93.6%. Classifying three weed species including Euphorbia maculata, Glechoma hederacea and Taraxacum officinale growing in perennial ryegrass was studied (Yu et al., 2019a) using VGGNet on RGB images. It achieved F-scores 98.6% and 95.6% for two independent test sets collected from two fields with different locations. A similar study was also conducted to classify three other species of weeds growing in perennial ryegrass including Hydrocotyle spp, Hedyotis cormybosa and Richardia scabra (Yu et al., 2019). Another study identified cephalanoplos, digitaria, bindweed and soybean in RGB images by introducing a CNN based on LeNet-5 (Ciresan et al., 2011) with a K-means clustering for unsupervised pre-training, which achieved an accuracy 92.9% (Tang et al., 2017). To further advance weed image classification in complex environments, a public dataset, namely DeepWeeds (Olsen et al., 2019), was constructed by acquiring RGB images in remote and extensive rangelands with rough and uneven terrains. The baseline accuracy 95.7% was achieved by a ResNet-50 for multi-label classification. A simplified DenseNet, namely DenseNet-128-32, was explored to reduce the computational cost and inference time, while keeping the performance comparable to that of the original DenseNet model (Lammie et al., 2019). More recent field-based studies have found image classification (including AlexNet, DenseNet, ResNet and VGGNet) outperformed object detection networks for the recognition of broadleaved seedlings in wheat (Zhuang et al., 2022). All image classification networks tested had F1 scores above 0.99.
Recently, a few fine-grained architectures were explored to improve weed image classification performance. By introducing graph-based image representation, a graph weeds net achieved the state-of-the-art accuracy 98.1% on the DeepWeeds dataset, which formulated global and fine-grained weed characteristics with GCNs (Hu et al., 2020). Another study also investigated the graph mechanisms (Jiang et al., 2020), in which GCN-ResNet-101 was proposed and the accuracy values varied from 96.5% to 98.9% on 4 public RGB datasets.
Deep unsupervised learning was explored in a recent weed study (dos Santos et al., 2019), which explored two methods: joint unsupervised learning of deep representations and image clusters (JULE) and deep clustering for unsupervised learning of visual features (DeepCluster). They adopted the CNN outputs as features for a clustering algorithm and specified pseudo labels for samples based on the clustering results. As reported, the DeepCluster method achieved an accuracy 70.6% with a VGG-16 backbone on the DeepWeeds dataset, and an accuracy 99.5% with an AlexNet backbone on Grass-Broadleaf.
Besides RGB images, multispectral imaging has also been investigated. Different sizes of multispectral images with a CNN involving 4 convolution layers were explored (Farooq et al., 2018). By varying the input size from \(125\times 125\) to \(500\times 500\), the accuracy varied from 86.3% to 94.7% on the UNSW Hyperspectral Weed Dataset. FCNN-SPLBP combined CNN and superpixel based local binary pattern feature extraction (Farooq et al., 2019), which was evaluated on two public datasets: an accuracy 89.7% for \(100\times 100\) pixel images on the UNSW Hyperspectral Weed Dataset and an accuracy 96.4% on the sugar beet/weed dataset. With the coarse, whole-image granularity of image classification, the method is more likely to be useful in coarse weed control scenarios, such as spot spraying (Calvert et al., 2021).
Weed object detection
Moving beyond the whole-image level of understanding, object detection provides bounding box coordinates of detected weeds. The additional contextual information allows targeting whole weed plants amongst crops, individual leaves, or other plant components. For instance, detecting individual leaves was found to be more effective than whole plant detection in strawberry raised beds (Sharpe et al., 2019). Existing weed object detection methods are mainly based on generic object detection methods. In (Dyrmann et al., 2017), DetectNet was used with an mIoU 64.0% and an F-score 60.3% on an in-house dataset containing 1,427 RGB images. Another study (Yu et al., 2019) with DetectNet achieved F-scores 99.8% and 100.0% for two different environments in detecting a weed species, namely Poa annua. In (Veeranampalayam Sivakumar et al., 2020), Faster R-CNN achieved an mIoU 84.0% and an F-score 67.0% in detecting waterhemp (Amaranthus tuberculatus), Palmer amaranth (Amaranthus palmeri), common lambsquarters (Chenopodiam album), velvetleaf (Abutilon theophrasti), and foxtail species such as yellow and green foxtails on an in-house dataset containing 450 augmented RGB images. YOLOv3 was used to detect broadleaves, sedges and grasses (Sharpe et al., 2020) and achieved an F-score 95.0%.
Weed semantic segmentation
With pixel-level granularity, semantic segmentation of weeds provides greater detail than object detection. The approach is more suited to precision weed control methods such as laser weeding, which must only hit the plant if they are to be effective. An intuitive approach for weed semantic segmentation is based on a two-stage scheme. Two CNNs were devised in (Potena et al., 2016), namely sNet and cNet, in which the first stage generated segmented objects and the second stage predicted the class of each object (Potena et al., 2016). The method were applied to multispectral images to identify the regions of crops, weeds and soil with a mean accuracy 92.0% and an mAP 97.4%. Another study adopted a conventional algorithm using HSV-colour room vegetation index method for segmentation and a CNN for classification (Knoll et al., 2018). It achieved an F-score 98.6 and a mean accuracy 97.9% on an in-house RGB image dataset containing carrots, weeds and soil.
FCNs were investigated in pursuit of end-to-end solutions which treat the segmentation and classification within one neural network. In (Dyrmann et al., 2016b; Mortensen et al., 2016), the last FC layer of a VGG-16 was replaced as a deconvolutional layer. The modified VGG-16 was evaluated on two RGB datasets: one to segment maize, weeds and soil and an mIoU 84.0% and a mean accuracy 95.4% was achieved; on another to segment equipment, soil, stump, weeds, grass, radish and unknown categories, a mean accuracy 79.0% was achieved. A FCN with a DenseNet backbone (Lottes et al., 2018) was evaluated to identify crops, weeds and soil in multispectral images and achieved F-Scores 86.6% and 92.4% on two datasets collected from sugar beet fields in two different locations. Two FCN-8 s were trained to segment RGB images: the first one recognized grass, clover, weeds and soil, and the latter one recognized fine-grained clover species including white clover and red clover (Skovsen et al., 2019). It achieved an mIoU 0.55 on its proposed GrassClover dataset.
In addition to using simple FCNs, recent studies tended to explore FCNs with additional mechanisms that are beneficial for segmentation tasks. In (Lameski et al., 2017), a SegNet with VGG-16 backbone achieved a mean accuracy 64.1% on a carrot-weed dataset containing RGB images of carrots, weeds and soil. In (Sa et al., 2017), a public multispectral dataset - Sugar beet/Weed - was proposed to identify crops, weeds and soil, and a SegNet with a VGG-16 backbone achieved an F-Score 80.0% and an AUC 78.0% by evaluating the crops and the weeds predictions within a binary pixel-wise classification scheme. In (Asad & Bais , 2019), a SegNet with a ResNet-50 backbone was adopted to identify canola and weeds in RGB images using a pre-processing step to remove backgrounds, which achieved an mIoU 82.9% and a mean accuracy 99.5%. A Bonnet framework (Milioto & Stachniss, 2019) to segment sunflower, weed and soil in RGB images achieved an mIoU 80.0% (Fawakherji et al., 2019b). A customized U-Net with different data augmentation strategies was investigated for the CWFID dataset (Brilhador et al., 2019), which achieved an F-score 83.4%. A VGG-UNet was evaluated for the Sugar beet/Weed dataset and achieved a mean accuracy 95.0% (Fawakherji et al., 2019a). DeepLab-v3 was evaluated on Sugarbeet2016 containing multispectral and depth data and an in-house RGB oilseed dataset, which achieved mIoU values of 87.1% and 88.9%, respectively (Wang et al., 2020).
Lightweight models aiming for efficient segmentation were explored. In (McCool et al., 2017), light models were mixed together with the guidance of a large and accurate model Inception-V3. On the CWFID dataset, compared to the Inception-V3 with an accuracy 93.9% and 0.12 fps during the inference, 4 mixed lightweight models achieved an accuracy 90.3% with 1.83 fps. A customized CNN using a ResNet-10 backbone (Li et al., 2019) with side outputs and short connections for multi-scale feature fusion achieved an F-score 98.0% and an mIoU 0.959 on the CWF-788.
Weed instance segmentation
Weed instance segmentation provides the highest-level granularity for weed recognition, with information on both pixel class and to which each weed the pixel belongs. As with semantic segmentation, the most likely use-case for the approach is with highly targeted weed control treatments. Understanding which weed to target rather than targeting every weed pixel, would greatly improve efficiency. There have been a few attempts at deploying instance segmentation algorithms for weed recognition. A recent study adopted Mask R-CNN for field RGB images of two crop species and four weed species (Champ et al., 2020). Further explorations for this approach are needed by considering the perspectives such as the improvements on different growth stage and small size plants.
Discussion
In this section, based on weed control challenges and the recent developments of deep learning techniques, we discuss the challenges and opportunities for further advancing weed recognition research from the following aspects: fine-grained learning, real-time inference, explainability, weakly-supervised/unsupervised learning, and incremental learning.
Fine-grained learning
As reviewed in “Weed recognition methods” section, most of the existing weed recognition methods were based on general deep architectures ignoring the challenge caused by the strong similarities of crops and weed species. Recently, 3 major categories of fine-grained deep methods were explored to address this challenge (Hu et al., 2019).
-
Patch-based methods based on the fact that the fine-grained details often occur at a local-level. With the patterns collected from each region, fused or aggregated methods can be used to compute the final outputs. For example, regional CNN based features can be collected according to the key points of human poses for fine-grained action recognition (Hu et al., 2019).
-
High-order pooling based methods were introduced to address fine-grained tasks as well, which did not require explicit patch proposals (e.g. (Zheng et al., 2019)). In particular, for a given convolutional feature map \({\textbf{X}}\in {\textbf{R}}^{c\times wh}\), the bilinear pooling can be computed by \({\textbf{X}}^\top {\textbf{X}}\); the trilinear pooling can be computed by \(({\textbf{X}}^\top {\textbf{X}}){\textbf{X}}\). The pooled vector can be used as the input of the subsequent layer of the network. The relations between the high-order pooling methods and the patch-based methods can be explained from the perspective of the attention (Kim et al., 2018). Both of these methods result in focusing on the critical regions to collect efficient deep representations for their associated tasks.
-
Regularization based methods are based on that the intra-class difference could be higher than the inter-class difference for fine-grained modelling, to introduce regularization terms for loss and drive the optimization to focus on learning fine-grained patterns. For example, in (Dubey et al., 2017), pair-wise confusion and entropic confusion was introduced to construct its loss function.
Such fine-grained deep models provide a great opportunity to advance weed recognition by taking the domain knowledge into account. For example, a weed can be decomposed into meaningful regions, such as leaves and stems. In our recent work, a patch-based GNN (Hu et al., 2020) was proposed towards fine-grained weed classification, which achieved an accuracy 98.1% on the DeepWeeds dataset, compared with the accuracy 95.3% of DenseNet.
Real-time inference
While most of the weed recognition studies demonstrated promising performance using deep learning techniques, these deep networks often contain a huge number of parameters. It leads to three major issues in regard to efficiency, memory consumption, and power consumption for deployment. Intuitively, as indicated in (Cheng et al., 2017), lightweight models (e.g. MobileNet (Howard et al., 2019)) can be devised by using mechanisms such as parameter pruning, low-rank factorization, transferred/compact convolutional filters. In particular, using a Google Pixel 3 device with one-thread on a single large core, the inference time of MobileNet (V3) for image classification on ImageNet achieved a top-1 accuracy 65.4 and inference latency 11.7 ms. Note that these lightweight models can also be treated as backbones for object detection and segmentation. For example, SSDLite with MobileNet (V3) Small backbone achieved an inference latency of 43 ms and an mAP 16.1 on COCO test set; MobileNet (V3) based segmentation achieved an mIoU 69.4 with an inference time of 1.03s for an input image with resolution of \(1024\times 2048\). For weed recognition, a ResNet-10 was proposed as backbone and introducing side outputs and short connections for multi-scale feature fusion. It achieved an mIoU 95.9 and an F-score 98.0, while the average inference latency is around 180 ms on an Nvidia Jeston TX2 (Li et al., 2019) (Table 5).
In addition to devising lightweight architectures, methods such as quantization and knowledge distillation are devised for any existing models with less parameters while providing comparable performance as the complex models (e.g. ResNet-50 vs. ResNet-152). Quantization methods reduce the number of bits to represent the parameters in a model. In particular, binarization only saves one bit for parameter, which significantly reduces the memory consumption and computational cost (Qin et al., 2020). A binarized DenseNet-128-32 was implemented by FPGA (Terasic DE1-SoC) for weed detection gaining an accuracy 88.91% (Lammie et al., 2019). It was slightly lower than a general DenseNet but obtained a very fast average inference latency 1.539 ms. Knowledge distillation follows a similar way in which human beings learn, which contains one or more large pre-trained teacher models and a small student model. It aims to obtain an efficient student model which mimics and performs comparably to the the teacher models. A distillation loss penalizes the difference between the outputs from the teacher and the student models. A weed recognition study followed this scheme to obtain a few lightweight models for semantic segmentation (McCool et al., 2017). Mixing these lightweight models achieved an accuracy 90.0% and the inference latency between 934ms to 546ms by using an Nvidia GeForce Titan X graphics card.
Weakly-supervised and unsupervised learning
As manually collected supervision information for weed dataset can be resource expensive, weakly-supervised and unsupervised learning algorithms are needed for weed recognition. For weakly-supervised learning, it is expected that weed object detection or even weed segmentation can be conducted when only using image-level annotation. For unsupervised learning, deep clustering and domain adaptation can be conducted. Deep clustering categorizes similar samples into one cluster in line with some similarity measures on their deep representations (Min et al., 2018). An application of deep clustering is that pre-training a neural network with a large unlabelled dataset and further fine-tuning on a small labelled dataset. Domain adaptation solves the problem that the training samples and testing samples following different distributions. This could be the case, for example, two datasets for the same species are from different locations. Therefore, unsupervised domain adaptation handles situations where a network is trained on labeled data from a source domain and unlabeled data from a related but different target domain. Readers can refer to (Wilson & Cook, 2020) for more details.
Note that existing deep learning based weed recognition methods have not adequately explored this realm to use the unlabelled samples. Until very recently, deep clustering was investigated for weed image classification (dos Santos et al., 2019), in which a VGG-16 based DeepCluster network achieved an accuracy 70.6% on the DeepWeeds dataset. In (Hu et al., 2020), a GraphWeedsNet involved a weakly-supervised learning strategy, namely multi-instance learning, and used image-level annotations to provide approximate locations of weed plants.
Explainable learning
Deep learning shows a black-box nature, since it is difficult to understand and interpret the relations between the inputs and the outputs. However, explainability is of great importance for building trust between models and users to eventually facilitate the model adoption. As summarized in (Xie et al., 2020b), there are three major approaches in pursuit of the explainability of deep learning: (1) visualization methods identify the most important parts of an input, which highly influence the results; (2) model distillation involves conventional machine learning models, most of which have clear statistical explanations and indications, to mimic the behavior of trained deep models; (3) intrinsic methods integrate mechanisms (e.g., the attention mechanisms).
Explainable learning has been seldom investigated for deep learning based weed recognition, although it has the potentials to provide further insights. Recently, graph weeds net was proposed with its graph mechanism, which treats the regions of an input image as graph vertices, to analyze the critical regions (Hu et al., 2020). As it usually takes more efforts for object detection or segmentation than image classification, such explainable learning approach also provides an opportunity to take less effort to focus on the critical objects within an image. Furthermore, the critical regions are obtained without regional annotations, which can be viewed as a weakly-supervised learning requiring less human efforts.
Incremental learning
Most of the existing weed recognition methods assume that a trained network will only deal with fixed target species, which are available during the training. As a result, when new species of interests are emerged, it is generally expected that the deep model needs to be re-trained with a new training set. To address this time-consuming and inflexible scheme, incremental learning is proposed to extends a trained model for new classes without the re-training from scratch. Note that the training samples of existing species are often not stored with a high volume due to storage limitations, whilst samples of incremental species could be adequate. Hence, incremental learning mainly addresses this imbalance nature when obtaining a new model based on an existing model.
To conduct incremental learning, 4 major approaches were investigated (De Lange et al., 2019). (1) retaining a subset of the old training data in line with a budget. (2) The distributions of the old dataset can be stored as the parameters of a generative model, which can produce unlimited samples during the incremental training. (3) Parameter isolation-based methods aim to prevent any possible forgetting of the previous tasks when no constraints on the model size. In general, for different species, it can use different model parameters to conduct the classification. (4) Regularization techniques prevent forgetting previous knowledge.
Recently, AgroAVNET explored the chance for incremental learning on the plant seedling dataset (Chavan & Nandedkar, 2018) and the accuracy achieved 91.35 for 12 species, compared to 93.64 from a general re-training. It followed a very straightforward way without fully exploiting the incremental learning, which froze the convolution layers trained by the original dataset and re-trained the FC layers only.
Large scale datasets
Large scale datasets are essential for developing high performance and robust deep learning models. For example, ImageNet (Krizhevsky et al., 2012), which contains 15 million labeled images belonging to roughly 22 000 categories, has played a significant role in advancing deep learning based vision tasks. However, as summarized in “Weed data” section, most of the existing weed datasets contain images of a small number of classes. In addition, those images were collected under limited scenarios, such as one growth stage and one light condition. This has limited the development of advanced methods applicable to a large variety of fields and prevents the translation towards commercial adoption. Therefore, constructing large scale datasets with diverse and complex conditions in the context of practical deployment can be highly demanded.
Conclusion
In this paper, we reviewed the recent progresses in the field of deep learning based weed recognition and discussed the challenges and opportunities for future research. After introducing the fundamentals of deep learning techniques, we provided a systematical review from three aspects: research data, evaluation, and weed recognition methods. There have been more than 10 public datasets collected through different modalities and many weed recognition methods have been reported across different research disciplines due to the inter-disciplinary nature of this topic. It is also noticed that most existing weed recognition methods were proposed by using the architectures developed for generic deep learning problems. Given the substantial differences in output granularity, the selection of specific recognition approaches should be governed by the in-field weed control treatment scenario. Where highly precise control methods are needed or where occlusion may reduce the effectiveness of coarser approaches, a trade-off may required in the complexity of architecture selected and hence complexity of training and deploying such an architecture in the field. Finally, we discussed the challenges and opportunities in terms of 5 different learning techniques and large scale dataset. Overall, deep learning based weed recognition has gained increasing interest from different research communities and we feel that large scale datasets are strongly needed to bring this research direction to a new level.
References
Asad, M. H., & Bais, A. (2019). Weed detection in canola fields using maximum likelihood classification and deep convolutional neural network. Information Processing in Agriculture, 7, 535.
Badrinarayanan, V., Kendall, A., & Cipolla, R. (2017). Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence, 39(12), 2481–2495.
Bah, M. D., Hafiane, A., & Canals, R. (2018). Deep learning with unsupervised data labeling for weed detection in line crops in uav images. Remote Sensing, 10(11), 1690.
Bawden, O., Kulk, J., Russell, R., McCool, C., English, A., Dayoub, F., Lehnert, C., & Perez, T. (2017). Robot for weed species plant-specific management. Journal of Field Robotics, 34(6), 1179–1199.
Bolya, D., Zhou, C., Xioa, F., & Lee, Y. J. (2019). Yolact: Real-time instance segmentation. In IEEE International Conference on Computer Vision (p. 9157). IEEE.
Brilhador, A., Gutoski, M., Hattori, L. T., de Souza, Inácio. A., Lazzaretti, A. E., & Lopes, H. S. (2019). Classification of weeds and crops at the pixel-level using convolutional neural networks and data augmentation. In IEEE Latin American conference on computational intelligence (pp. 1–6). IEEE.
Bruna, J., Zaremba, W., Szlam, A., & LeCun, Y. (2013). Spectral networks and locally connected networks on graphs. Preprint retrieved from http://arxiv.org/abs/1312.6203
Calvert, B., Olsen, A., Whinney, J., & Rahimi Azghadi, M. (2021). Robotic spot spraying of Harrisia cactus (Harrisia martinii) in grazing pastures of the australian rangelands. Plants, 10(10), 2054.
Champ, J., Mora-Fallas, A., Goëau, H., Mata-Montero, E., Bonnet, P., & Joly, A. (2020). Instance segmentation for the fine detection of crop and weed plants by precision agricultural robots. Applications in Plant Sciences, 8(7), e11373.
Charters, J., Wang, Z., Chi, Z., Tsoi, A. C., & Feng, D. D. (2014). Eagle: A novel descriptor for identifying plant species using leaf lamina vascular features. In IEEE international conference on multimedia and expo workshops (pp. 1–6). IEEE.
Chavan, T. R., & Nandedkar, A. V. (2018). Agroavnet for crops and weeds classification: A step forward in automatic farming. Computers and Electronics in Agriculture, 154, 361–372.
Chebrolu, N., Lottes, P., Schaefer, A., Winterhalter, W., Burgard, W., & Stachniss, C. (2017). Agricultural robot dataset for plant classification, localization and mapping on sugar beet fields. The International Journal of Robotics Research, 36(10), 1045–1052.
Chen, L. C., Zhu, Y., Papandreou, G., Schroff, F., & Adam, H. (2018). Encoder-decoder with atrous separable convolution for semantic image segmentation. In European conference on computer vision (pp. 801–818). Springer.
Chen, H., Sun, K., Tian, Z., Shen, C., Huang, Y., & Yan, Y. (2020). Blendmask: Top-down meets bottom-up for instance segmentation. In IEEE conference on computer vision and pattern recognition (pp 8573–8581)
Cheng, Y., Wang, D., Zhou, P., & Zhang, T. (2017). A survey of model compression and acceleration for deep neural networks. Preprint retrieved from http://arxiv.org/abs/1710.09282
Ciresan, D. C., Meier, U., Gambardella, L. M., & Schmidhuber, J. (2011). Convolutional neural network committees for handwritten character classification. In International conference on document analysis and recognition (pp 1135–1139). IEEE.
Coleman, G. R. Y., Stead, A., Rigter, M. P., Xu, Z., Johnson, D., Brooker, G. M., Sukkarieh, S., & Walsh, M. J. (2019). Using energy requirements to compare the suitability of alternative methods for broadcast and site-specific weed control. Weed Technology, 33(4), 633–650.
Cortes, C., & Vapnik, V. (1995). Support-vector networks. Machine Learning, 20(3), 273–297.
De Lange, M., Aljundi, R., Masana, M., Parisot, S., Jia, X., Leonardis, A., Slabaugh, G., & Tuytelaars, T. (2019). Continual learning: A comparative study on how to defy forgetting in classification tasks. Preprint retrieved form http://arxiv.org/abs/1909.08383
dos Santos, F. A., Freitas, D. M., da Silva, G. G., Pistori, H., & Folhes, M. T. (2017). Weed detection in soybean crops using convnets. Computers and Electronics in Agriculture, 143, 314–324.
dos Santos, F. A., Freitas, D. M., da Silva, G. G., Pistori, H., & Folhes, M. T. (2019). Unsupervised deep learning and semi-automatic data labeling in weed discrimination. Computers and Electronics in Agriculture, 165, 104963.
Dubey, A., Gupta, O., Guo, P., Raskar, R., Farrell, R., & Naik, N. (2017). Training with confusion for fine-grained visual classification. Preprint retrieved from http://arxiv.org/abs/1705.08016
Dyrmann, M., Jørgensen, R. N., & Midtiby, H. S. (2017). Roboweedsupport-detection of weed locations in leaf occluded cereal crops using a fully convolutional neural network. Advances in Animal Bioscience, 8(2), 842–847.
Dyrmann, M., Karstoft, H., & Midtiby, H. S. (2016a). Plant species classification using deep convolutional neural network. Biosystems Engineering, 151, 72–80.
Dyrmann, M., Mortensen, A. K., Midtiby, H. S., & Jørgensen, R. N. (2016b). Pixel-wise classification of weeds and crops in images by using a fully convolutional neural network. In International conference on agricultural engineering (pp 26–29).
Farooq, A., Hu, J., & Jia, X. (2018). Analysis of spectral bands and spatial resolutions for weed classification via deep convolutional neural network. IEEE Geoscience and Remote Sensing Letters, 16(2), 183–187.
Farooq, A., Jia, X., Hu, J., & Zhou, J. (2019). Multi-resolution weed classification via convolutional neural network and superpixel based local binary pattern using remote sensing images. Remote Sensing, 11(14), 1692.
Fawakherji, M., Potena, C., Bloisi, D. D., Imperoli, M., Pretto, A., & Nardi, D. (2019a). Uav image based crop and weed distribution estimation on embedded gpu boards. In International conference on computer analysis of images and patterns (pp 100–108). Springer.
Fawakherji, M., Youssef, A., Bloisi, D., Pretto, A., & Nardi, D. (2019b). Crop and weeds classification for precision agriculture using context-independent pixel-wise segmentation. In IEEE international conference on robotic computing (pp 146–152). IEEE.
Gai, J., Tang, L., & Steward, B. L. (2020). Automated crop plant detection based on the fusion of color and depth images for robotic weed control. Journal of Field Robotics, 37(1), 35–52.
Garg, V. K., Jegelka, S., & Jaakkola, T. (2020). Generalization and representational limits of graph neural networks. Preprint retrieved from http://arxiv.org/abs/2002.06157
Gharde, Y., Singh, P., Dubey, R., & Gupta, P. (2018). Assessment of yield and economic losses in agriculture due to weeds in India. Crop Protection, 107, 12–18.
Giselsson, T. M., Jørgensen, R. N., Jensen, P. K., Dyrmann, M., & Midtiby, H. S. (2017). A public image database for benchmark of plant seedling classification algorithms. Preprint retrieved from http://arxiv.org/abs/1711.05458
Gu, J., Wang, Z., Kuen, J., Ma, L., Shahroudy, A., Shuai, B., Liu, T., Wang, X., Wang, G., Cai, J., & Chen, T. (2018). Recent advances in convolutional neural networks. Pattern Recognition, 77, 354–377.
Haug, S., & Ostermann, J. (2014). A crop/weed field image dataset for the evaluation of computer vision based precision agriculture tasks. In European conference on computer vision (pp 105–116). Springer.
He, K., Gkioxari, G., Dollár, P., & Girshick, R. (2017). Mask R-CNN. In IEEE international conference on computer vision (pp 2961–2969).
Howard, A., Sandler, M., Chu, G., Chen, L. C., Chen, B., Tan, M., Wang, W., Zhu, Y., Pang, R., & Vasudevan, V. (2019). Searching for mobilenetv3. In IEEE international conference on computer vision (pp 1314–1324).
Huang, G., Liu, Z., Maaten, L., & Weinberger, K. Q. (2017). Densely connected convolutional networks. In IEEE conference on computer vision and pattern recognition (pp 2261–2269). https://doi.org/10.1109/CVPR.2017.243
Hu, K., Coleman, G., Zeng, S., Wang, Z., & Walsh, M. (2020). Graph weeds net: A graph-based deep learning method for weed recognition. Computers and Electronics in Agriculture, 174, 105520.
Hu, K., Wang, Z., Mei, S., Ehgoetz Martens, K. A., Yao, T., Lewis, S. J. G., & Feng, D. D. (2019). Vision-based freezing of gait detection with anatomic directed graph representation. IEEE Journal of Biomedical and Health Informatics, 24(4), 1215–1225.
Jiang, H., Zhang, C., Qiao, Y., Zhang, Z., Zhang, W., & Song, C. (2020). CNN feature based graph convolutional network for weed and crop recognition in smart farming. Computers and Electronics in Agriculture, 174, 105450.
Kim, J. H., Jun, J., & Zhang, B. T. (2018). Bilinear attention networks. In Advances in neural information processing systems (pp 1564–1574).
Knoll, F. J., Czymmek, V., Poczihoski, S., Holtorf, T., & Hussmann, S. (2018). Improving efficiency of organic farming by using a deep learning classification approach. Computers and Electronics in Agriculture, 153, 347–356.
Krizhevsky, A., Sutskever, I., Hinton, G. E. (2012). ImageNet classification with deep convolutional neural networks. Advances in neural information processing systems (pp 1097–1105).
Lameski, P., Zdravevski, E., Trajkovik, V., Kulakov, A. (2017). Weed detection dataset with rgb images taken under variable light conditions. In International conference on ICT innovations (pp 112–119). Springer.
Lammie, C., Olsen, A., Carrick, T., & Azghadi, M. R. (2019). Low-power and high-speed deep fpga inference engines for weed classification at the edge. IEEE Access, 7, 51171–51184.
Le Nguyen Thanh, V., Apopei, B., & Alameh, K. (2019). Effective plant discrimination based on the combination of local binary pattern operators and multiclass support vector machine methods. Information Processing in Agriculture, 6(1), 116–131. https://doi.org/10.1016/j.inpa.2018.08.002
Lin, T. Y., Goyal, P., Girshick, R., He, K., Dollár, P. (2017). Focal loss for dense object detection. In IEEE international conference on computer vision (pp 2980–2988). IEEE.
Li, Y., Qi, H., Dai, J., Ji, X., Wei, Y. (2017). Fully convolutional instance-aware semantic segmentation. In IEEE conference on computer vision and pattern recognition (pp 2359–2367). IEEE.
Liu, W., Anguelov, D., Erhan, D., Szegedy, C., Reed, S., Fu, C. Y., Berg, A. C. (2016). Ssd: Single shot multibox detector. In European conference on computer vision (pp 21–37). Springer.
Li, N., Zhang, X., Zhang, C., Guo, H., Sun, Z., & Wu, X. (2019). Real-time crop recognition in transplanted fields with prominent weed growth: a visual-attention-based approach. IEEE Access, 7, 185310–185321.
Llewellyn, R., Ronning, D., Clarke, M., Mayfield, A., Walker, S., & Ouzman, J. (2016). Impact of weeds in Australian grain production. Grains Research and Development, 2016, 1–10.
Long, J., Shelhamer, E., Darrell, T. (2015). Fully convolutional networks for semantic segmentation. In IEEE conference on computer vision and pattern recognition (pp 3431–3440). IEEE.
Lottes, P., Behley, J., Milioto, A., & Stachniss, C. (2018). Fully convolutional networks with sequential information for robust crop and weed detection in precision farming. IEEE Robotics and Automation Letters, 3(4), 2870–2877.
Machleb, J., Peteinatos, G. G., Kollenda, B. L., Andújar, D., & Gerhards, R. (2020). Sensor-based mechanical weed control: Present state and prospects. Computers and Electronics in Agriculture, 176, 105638.
McCool, C., Perez, T., & Upcroft, B. (2017). Mixtures of lightweight deep convolutional neural networks: Applied to agricultural robotics. IEEE Robotics and Automation Letters, 2(3), 1344–1351.
Milioto, A., & Stachniss, C. (2019). Bonnet: An open-source training and deployment framework for semantic segmentation in robotics using CNNs. In International conference on robotics and automation (pp 7904–7100). IEEE.
Min, E., Guo, X., Liu, Q., Zhang, G., Cui, J., & Long, J. (2018). A survey of clustering with deep learning: From the perspective of network architecture. IEEE Access, 6, 39501–39514.
Mortensen, A. K., Dyrmann, M., Karstoft, H., Jørgensen, R. N., & Gislum, R. (2016). Semantic segmentation of mixed crops using deep convolutional neural network. In International conference of agricultural engineering (pp 26–29).
Olsen, A., Konovalov, D. A., Philippa, B., Ridd, P., Wood, J. C., Johns, J., Banks, W., Girgenti, B., Kenny, O., Whinney, J., et al. (2019). Deepweeds: A multiclass weed species image dataset for deep learning. Scientific Reports, 9(1), 2058.
Papandreou, G., Zhu, T., Chen, L. C., Gidaris, S., Tompson, J., Murphy, K. (2018). Personlab: person pose estimation and instance segmentation with a bottom-up, part-based, geometric embedding model. In European Conference on Computer Vision (pp 269–286). Springer.
Picon, A., San-Emeterio, M. G., Bereciartua-Perez, A., Klukas, C., Eggers, T., & Navarra-Mestre, R. (2022). Deep learning-based segmentation of multiple species of weeds and corn crop using synthetic and real image datasets. Computers and Electronics in Agriculture, 194, 106719.
Potena, C., Nardi, D., Pretto, A. (2016). Fast and accurate crop and weed identification with summarized train sets for precision agriculture. In International conference on intelligent autonomous systems (pp 105–121). Springer.
Qin, H., Gong, R., Liu, X., Bai, X., Song, J., Sebe, N. (2020). Binary neural networks: A survey. Pattern Recognition, 2020, 107281.
Redmon, J., Divvala, S., Girshick, R., Farhadi, A. (2016). You only look once: Unified, real-time object detection. In IEEE conference on computer vision and pattern recognition (pp 779–788)
Ren, S., He, K., Girshick, R., Sun, J. (2015). Faster R-CNN: Towards real-time object detection with region proposal networks. In Advances in neural information processing systems (pp 91–99).
Ronneberger, O., Fischer, P., & Brox, T. (2015). U-net: Convolutional networks for biomedical image segmentation. In International conference on medical image computing and computer-assisted intervention (pp. 234–241). Springer.
Sa, I., Chen, Z., Popović, M., Khanna, R., Liebisch, F., Nieto, J., & Siegwart, R. (2017). weednet: Dense semantic weed classification using multispectral images and mav for smart farming. IEEE Robotics and Automation Letters, 3(1), 588–595.
Salazar-Gomez, A., Darbyshire, M., Gao, J., Sklar, E. I., Parsons, S. (2021). Towards practical object detection for weed spraying in precision agriculture. Preprint retrieved from http://arxiv.org/abs/2109.11048
Schmidhuber, J. (2015). Deep learning in neural networks: An overview. Neural Networks, 61, 85–117.
Sharpe, S. M., Schumann, A. W., & Boyd, N. S. (2019). Detection of Carolina geranium (Geranium carolinianum) growing in competition with strawberry using convolutional neural networks. Weed Science, 67(2), 239–245.
Sharpe, S. M., Schumann, A. W., Yu, J., & Boyd, N. S. (2020). Vegetation detection and discrimination within vegetable plasticulture row-middles using a convolutional neural network. Precision Agriculture, 21(2), 264–277.
Shorten, C., & Khoshgoftaar, T. M. (2019). A survey on image data augmentation for deep learning. Journal of Big Data, 6(1), 60.
Simonyan, K., & Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. Preprint retrieved from http://arxiv.org/abs/1409.1556
Skovsen, S., Dyrmann, M., Mortensen, A. K., Laursen, M. S., Gislum, R., Eriksen, J., Farkhani, S., Karstoft, H., & Jorgensen, R. N. (2019). The grassclover image dataset for semantic and hierarchical species understanding in agriculture. In IEEE conference on computer vision and pattern recognition workshops. IEEE.
Szegedy, C., Ioffe, S., Vanhoucke, V., Alemi, A. A. (2017). Inception-v4, inception-resnet and the impact of residual connections on learning. In AAAI conference on artificial intelligence. IEEE.
Tang, J., Wang, D., Zhang, Z., He, L., Xin, J., & Xu, Y. (2017). Weed identification based on k-means feature learning combined with convolutional neural network. Computers and Electronics in Agriculture, 135, 63–70.
Tian, Z., Shen, C., Chen, H., & He, T. (2019). Fcos: Fully convolutional one-stage object detection. IEEE international conference on computer vision (pp 9627–9636). IEEE.
Veeranampalayam Sivakumar, A. N. V., Li, J., Scott, S., Psota, E., Jhala, J. A., Luck, J. D., & Shi, Y. (2020). Comparison of object detection and patch-based classification deep learning models on mid-to late-season weed detection in uav imagery. Remote Sensing, 12(13), 2136.
Veličković, P., Cucurull, G., Casanova, A., Romero, A., Liò, P., & Bengio, Y. (2018). Graph attention networks. International conference on learning representations, https://openreview.net/forum?id=rJXMpikCZ
Wang, A., Xu, Y., Wei, X., & Cui, B. (2020). Semantic segmentation of crop and weed using an encoder-decoder network and image enhancement method under uncontrolled outdoor illumination. IEEE Access, 8, 81724–81734.
Wilson, G., & Cook, D. J. (2020). A survey of unsupervised deep domain adaptation. ACM Transactions on Intelligent Systems and Technology, 11(5), 1–46.
Xie, E., Sun, P., Song, X., Wang, W., Liu, X., Liang, D., Shen, C., & Luo, P. (2020a). Polarmask: Single shot instance segmentation with polar representation. In IEEE conference on computer vision and pattern recognition (pp 12193–12202). IEEE.
Xie, N., Ras, G., van Gerven, M., Doran, D. (2020b). Explainable deep learning: A field guide for the uninitiated. Preprint retrieved from http://arxiv.org/abs/2004.14545
Xinshao, W., & Cheng, C. (2015). Weed seeds classification based on pcanet deep learning baseline. In Asia-Pacific signal and information processing association annual summit and conference (pp. 408–415). IEEE.
Yu, J., Schumann, A. W., Cao, Z., Sharpe, S. M., & Boyd, N. S. (2019a). Weed detection in perennial ryegrass with deep learning convolutional neural network. Frontiers in Plant Science, 2019, 10.
Yu, J., Sharpe, S. M., Schumann, A. W., & Boyd, N. S. (2019b). Deep learning for image-based weed detection in turfgrass. European Journal of Agronomy, 104, 78–84.
Zeiler, M. D., Krishnan, D., Taylor, G. W., & Fergus, R. (2010). Deconvolutional networks. In IEEE conference on computer vision and pattern recognition (pp. 2528–2535). IEEE.
Zhao, H., Shi, J., Qi, X., Wang, X., & Jia, J. (2017). Pyramid scene parsing network. In IEEE conference on computer vision and pattern recognition (pp. 2881–2890). IEEE.
Zheng, H., Fu, J., Zha, Z. J., & Luo, J. (2019). Looking for the devil in the details: Learning trilinear attention sampling network for fine-grained image recognition. In IEEE conference on computer vision and pattern recognition (pp. 5012–5021). IEEE.
Zhuang, J., Li, X., Bagavathiannan, M., Jin, X., Yang, J., Meng, W., Li, T., Li, L., Wang, Y., Chen, Y., & Yu, J. (2022). Evaluation of different deep convolutional neural networks for detection of broadleaf weed seedlings in wheat. Pest Management Science, 78(2), 521–529.
Zoph, B., Vasudevan, V., Shlens, J., & Le, Q. V. (2018). Learning transferable architectures for scalable image recognition. In IEEE conference on computer vision and pattern recognition (pp. 8697–8710). IEEE.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This work was supported by the GRDC (Grains Research and Development Corporation) [Grant Number 9177493].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hu, K., Wang, Z., Coleman, G. et al. Deep learning techniques for in-crop weed recognition in large-scale grain production systems: a review. Precision Agric 25, 1–29 (2024). https://doi.org/10.1007/s11119-023-10073-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11119-023-10073-1