Abstract
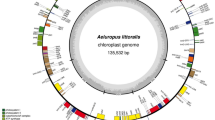
Adinandra bockiana E. Pritz. ex Diels is a potential plant of the Adinandra genus that contains bioactive. This study presents the results of assembly and annotation of the chloroplast (cp) genome of A. bockiana, a comparison of it with Adinandra cp genomes of several other species, and proposed candidate DNA barcoding from the cp genome. The A. bockiana cp genome was reconstructed by raw reads from total DNA, assembling reads (each read is 150 bp in length) into contigs, and then the contigs were assembled again to sequence the complete cp genome. The A. bockiana cp genome is 156,284 bp in length and consists of a large single-copy (LSC) region of 85,693 bp, a small single-copy (SSC) region of 18,411 bp, a pair of inverted repeats (IRa and IRb) of 26,090 bp each, 129 functional genes, 51 SSRs, and other 70 repeat structures. The functional genes of the A. bockiana cp genome contain 84 protein-coding genes, 37 transfer RNA genes, and eight ribosomal RNA genes. A comparative analysis of the A. bockiana cp genome with the rest of the public Adinandra genomes showed that the number of genes and their structure were highly conserved in the surveyed group. Based on complete cp genomes and matK and trnL sequences of some species of the Adinandra and other genera, molecular phylogenetic trees were established which show the genetic relationships of the species belonging to the Pentaphylacaceae family. Among the chloroplast genes of A. bockiana, the matK and trnL genes are better candidates for phylogenetic analysis.





Similar content being viewed by others
References
Amiryousefi A, Hyvönen J, Poczai P (2018) IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34:3030–3031
Beier S, Thiel T, Münch T, Scholz U, Mascher M (2017) MISA-web: a web server for microsatellite prediction. Bioinformatics 33:2583–2585
Bendich AJ (2004) Circular chloroplast chromosomes: the grand illusion. Plant Cell 16:1661–1666
Brad K, Zhang Y (2018) Study on extraction and purification of apigenin and the physical and chemical properties of its complex with lecithin. Pharmacogn Mag 14:203–206
Chen Y, Chen G, Fu X, Liu RH (2015) Phytochemical profiles and antioxidant activity of different varieties of Adinandra tea (Adinandra Jack). J Agric Food Chem 63:169–176
Chen Y, She G, Chen H (1997) Study on antitumor activities of substances extracted from Adinandra nitida Merr. ex H. Li. J Shantou Univ, Nat Sci 12:43–45
Cheng Y, Zhang L, Qi J, Zhang L (2020) Complete chloroplast genome sequence of Hibiscus cannabinus and comparative analysis of the Malvaceae family. Front Genet 11:227
Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J (2013) Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569
Daniell H, Lin CS, Yu M, Chang WJ (2016) Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol 17:134
Dong W, Cheng T, Li C, Xu C, Long P, Chen C, Zhou S (2014) Discriminating plants using the DNA barcode rbcLb: an appraisal based on a large data set. Mol Ecol Resour 14:336–343
Dong W, Liu J, Yu J, Wang L, Zhou S (2012) Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 7:e35071
Gao H, Liu B, Liu F, Chen Y (2010) Anti-proliferative effect of camellianin A in Adinandra nitida leaves and its apoptotic induction in human Hep G2 and MCF-7 cells. Molecules (basel, Switzerland) 15:3878–3886
Gao X, Zhang X, Meng H, Li J, Zhang D, Liu C (2018) Comparative chloroplast genomes of Paris Sect. Marmorata: insights into repeat regions and evolutionary implications. BMC Genomics 19:878
He S, Wang Y, Volis S, Li D, Yi T (2012) Genetic diversity and population structure: implications for conservation of wild soybean (Glycine soja Sieb. et Zucc) based on nuclear and chloroplast microsatellite variation. Int J Mol Sci 13:12608–12628
Ho PH (1999) Cay Co Vietnam (an illustrated flora of Vietnam). Youth Publishing House, Ho Chi Minh (in Vietnamese)
Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee S-B, Peery R, McNeal JR, Kuehl JV, Boore JL (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci 104:19369
Jansen RK, Ruhlman TA (2012) Plastid genomes of seed plants. In: Knoop V (ed) Bock R. Springer, Genomics of chloroplasts and mitochondria Advances in Photosynthesis and Respiration (Including Bioenergy and Related Processes) Dordrecht, pp 103–126
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9
Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166
Kawata M, Harada T, Shimamoto Y, Oono K, Takaiwa F (1997) Short inverted repeats function as hotspots of intermolecular recombination giving rise to oligomers of deleted plastid DNAs (ptDNAs). Curr Genet 31:179–184
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Kurtz S, Schleiermacher C (1999) REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics 15:426–427
Liang H, Zhang Y, Deng J, Gao G, Ding C, Zhang L, Yang R (2020) The complete chloroplast genome sequences of 14 Curcuma species: insights into genome evolution and phylogenetic relationships within Zingiberales. Front Genet 11:802
Liu B, Yang J, Ma Y, Yuan E, Chen C (2010) Antioxidant and angiotensin converting enzyme (ACE) inhibitory activities of ethanol extract and pure flavonoids from Adinandra nitida leaves. Pharm Biol 48:1432–1438
Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X (2012) CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics 13:715
Lohse M, Drechsel O, Bock R (2007) OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52:267–274
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046–1047
Medgyesy P, Fejes E, Maliga P (1985) Interspecific chloroplast recombination in a nicotiana somatic hybrid. Proc Natl Acad Sci USA 82:6960–6964
Min TL, Bruce BB (2007a) Theaceae. In: Wu Z. Y. RPHaHDY (ed) Flora of China 12. Science Press, Beijing and Missouri Botanical Garden Press, St. Louis, pp 435–443
Min TL, Bruce BB (2007b) Theaceae. In: Wu Z. Y., Raven P. H. and Hong D. Y. (eds.), Flora of China 12:435–443. Science Press, Beijing and Missouri Botanical Garden Press, St. Louis
Mustárdy L, Buttle K, Steinbach G, Garab G (2008) The three-dimensional network of the thylakoid membranes in plants: quasihelical model of the granum-stroma assembly. Plant Cell 20:2552
Nguyen L, Nguyen Q, Nguyen N, Vi T, Sy T, Nguyen T, Chu M (2020) Antibacterial, antioxidant and anti - cancerous activities of Adiandra megaphylla Hu leaf extracts. Biosci Biotechnol Res Commun 13:1015–1020
Nguyen HQ, Nguyen TNL, Doan TN, Nguyen TTN, Pham MH, Le TL, Sy DT, Chu HH, Chu HM (2021a) Complete chloroplast genome of novel Adinandra megaphylla Hu species: molecular structure, comparative and phylogenetic analysis. Sci Rep 11:11731. https://doi.org/10.1038/s41598-021-91071-z
Nguyen QH, Nguyen LTN, Nguyen NTT, Sy TD, Chu MH,Chu HH, Le LT, Doan NT, Pham HM, Doan GTH (2021b) Adinandra bockiana chloroplast, complete genome. GenBank MW699853.1
Nguyen TB (2003) A list of all plant species in Vietnam. Agriculture Publish House, Hanoi
Okonechnikov K, Golosova O, Fursov M, Ugene Team (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167
Palmer JD (1985) Comparative organization of chloroplast genomes. Annu Rev Genet 19:325–354
Rosea JP, Kleistb TJ, Löfstrandc SD, Drewd BT, Schönenbergere J, Sytsma KJ (2018) Phylogeny, historical biogeography, and diversification of angiosperm order Ericales suggest ancient Neotropical and East Asian connections. Mol Phylogenet Evol 122:59–79
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302
Schattat M, Barton K, Baudisch B, Klösgen RB, Mathur J (2011) Plastid stromule branching coincides with contiguous endoplasmic reticulum dynamics. Plant Physiol 155:1667–1677
Shaw J, Shafer HL, Leonard OR, Kovach MJ, Schorr M, Morris AB (2014) Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. Am J Bot 101:1987–2004
Souza HA, Muller LA, Brandão RL, Lovato MB (2012) Isolation of high quality and polysaccharide-free DNA from leaves of Dimorphandra mollis (Leguminosae), a tree from the Brazilian Cerrado. Genet Mol Res 11:756–764
Souza UJBd, Nunes R, Targueta CP, Diniz-Filho JAF, Telles MPdC (2019) The complete chloroplast genome of Stryphnodendron adstringens (Leguminosae - Caesalpinioideae): comparative analysis with related mimosoid species. Sci Rep 9:14206
Steven PF (2017) Angiosperm phylogeny. In: http://www.mobot.org/MOBOT/research/APweb/
Sullivan AR, Schiffthaler B, Thompson SL, Street NR, Wang XR (2017) Interspecific plastome recombination reflects ancient reticulate evolution in Picea (Pinaceae). Mol Biol Evol 34:1689–1701
Wang X, Cheng F, Rohlsen D, Bi C, Wang C, Xu Y, Wei S, Ye Q, Yin T, Ye N (2018) Organellar genome assembly methods and comparative analysis of horticultural plants. Horticulture Res 2018:5
Wu CC, Hsu ZF, Tsou CH (2007a) Phylogeny and taxonomy of Eurya (Ternstroemiaceae) from Taiwan, as inferred from ITS sequence data. Bot Stud 48:97–116
Wu Z, Raven PH, Hong De (2007b) Flora of China. Science Press, Beijing, and Missouri Botanical Garden Press, St. Louis
Yu J, Chen M (1997) Studies on flavonoids extraction from Adinandra nitida Merr. ex H. L. Li. and on their antioxidative and bacteriostatic bioactivities. J Shantou Univ Nat Sci 12:52–56
Yu XQ, Gao LM, Soltis DE, Soltis PS, Yang JB, Fang L, Yang SX, Li DZ (2017) Insights into the historical assembly of East Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytol 215:1235–1248
Yuan C, Huang L, Suh JH, Wang Y (2019) Bioactivity-guided isolation and identification of antiadipogenic compounds in Shiya tea (Leaves of Adinandra nitida). J Agric Food Chem 67:6785–6791
Zhang Y, Du L, Liu A, Chen J, Wu L, Hu W, Zhang W, Kim K, Lee SC, Yang TJ, Wang Y (2016) The complete chloroplast genome sequences of five epimedium species: lights into phylogenetic and taxonomic analyses. Front Plant Sci 7:306
Zuo W, Xu J, Zhou C, Gan L (2010) [Simultaneous determination of five flavonoid compounds in leaves of Adinandra nitida by HPLC-PAD]. Zhongguo zhongyao zazhi = China Journal of Chinese Materia Medica 35:2406–2409
Funding
This research is funded by Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 106.02-2018.338; Vietnam Ministry of Education and Training under grant number B2022-TNA-43.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
• The complete chloroplast genome of Adinandra bockiana was sequenced, and its size is 156,284 bp, including an LSC, SSC, 129 functional genes, 51 SSRs, and 70 repeat structures.
• The phylogenetic reconstruction using comparative analysis with species of the Pentaphylacaceae family and the matK sequence is a better candidate for phylogenetic resolution.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nguyen, N.T.T., Pho, H.T.T., Nguyen, Q.H. et al. Characteristics of the Chloroplast Genome of Adinandra bockiana and Comparative Analysis with Species of Pentaphylacaceae Family. Plant Mol Biol Rep 41, 611–621 (2023). https://doi.org/10.1007/s11105-023-01389-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-023-01389-3