Abstract
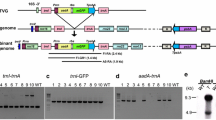
We have determined the nucleotide sequences around the junction points of oligomeric-deleted ptDNAs possessing a head-to-head or tail-to-tail configuration from long-term cultured cell lines and albino plants. It was shown that DNA rearrangement occurred by direct fusion of deleted ptDNAs in an inverted orientation, which was linked by an asymmetrical sequence of 254–698 bp derived from either of the ptDNAs joined. It is notable that inverted repeats of 7–14 bp flank the asymmetrical sequences at each of the junction points. These features of the DNA sequence around the junction points are commonly observed in oligomeric ptDNA with a large-scale deletion regardless of the cell lines employed. It is suggested that the short inverted repeats are involved in the intermolecular recombination of ptDNA.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 1 July / 21 October 1996
Rights and permissions
About this article
Cite this article
Kawata, M., Harada, T., Shimamoto, Y. et al. Short inverted repeats function as hotspots of intermolecular recombination giving rise to oligomers of deleted plastid DNAs (ptDNAs). Curr Genet 31, 179–184 (1997). https://doi.org/10.1007/s002940050193
Issue Date:
DOI: https://doi.org/10.1007/s002940050193