Abstract
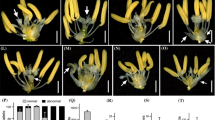
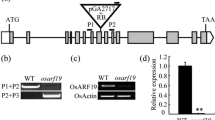
Floral organ identity and specific number directly affect anthesis habits, fertilization and grain yield. Here, we identified a deformed interior floral organ 1 (difo1) mutant from selfing progenies of indica cv. Zhonghui8015 (Zh8015) after 60Co γ-ray treatment. Compared with the Zh8015 spikelet, the interior floral organs of the difo1 mutant present various numbers of stamens and stigmas, with no typical filament and no mature pollen grains. Most difo1 flowers exhibited an increased number of stigmas that were attached to the stamens and an intumescent ovule-like cell mass in addition to the ovary. Transverse sections of spikelets and scanning electron microscopy analysis revealed an indeterminate number of interior floral organs and abnormal early spikelet development for the difo1 mutant. Instead of the linear-shaped surface of wild-type stamens, difo1 displayed a glossy stamen surface resulting in immature stamens and complete sterility. In addition, the difo1 mutant exhibited delayed anthesis, rapid anthesis and non-extended stamens compared with wild type. Genetic analysis and gene mapping revealed that difo1 was controlled by a single recessive gene, which was fine-mapped to a 54-kb interval on the short arm of chromosome 4 between markers S22 and RM16439 harboring nine ORFs. Sequence analysis revealed that the mutant carried a single nucleotide deletion in its promoter region, which likely corresponded to the phenotype, in a C2H2-type zinc finger protein gene (LOC_Os04g08600). Moreover, qRT-PCR analysis showed a significantly down-regulated expression pattern for DIFO1 and many floral organ identity genes in the interior floral organs of difo1. DIFO1 is therefore an important floral organ development gene in rice, particularly with regard to interior organ meristem identity and floret primordium differentiation.






Similar content being viewed by others
Abbreviations
- CTAB:
-
Cetyltrimethyl ammonium bromide
- InDel:
-
Insertion/deletion
- kb:
-
Kilobase
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
- qRT-PCR:
-
Quantitative real-time PCR
- SEM:
-
Scanning electron microscope
- SSR:
-
Simple sequence repeats
References
Breuil-Broyer S, Trehin C, Morel P, Boltz V, Sun B, Chambrier P, Ito T, Negrutiu I (2016) Analysis of the Arabidopsis superman allelic series and the interactions with other genes demonstratedevelopmental robustness and joint specification of male–female boundary, flower meristem termination andcarpel compartmentalization. Ann Bot 117:905–923
Byzova MV, Franken J, Aarts MG, de Almeida-Engler J, Engler G, Mariani C, Van Lookeren Campagne MM, Angenent GC (1999) Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes Dev 13:1002–1014
Chen Q, Atkinson A, Otsuga D, Christensen T, Reynolds L, Drews GN (1999) The Arabidopsis FILAMENTOUS FLOWER gene is required for flower formation. Development 126:2715–2726
Chu H, Qian Q, Liang W, Yin C, Tan H, Yao X, Yuan Z, Yang J, Huang H, Luo D, Ma H, Zhang D (2006) The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol 142:1039–1052
Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353:31–37
Colombo L, Franken J, Koetje E, van Went J, Dons HJ, Angenent GC, van Tunen AJ (1995) The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7:1859–1868
Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z (2010) Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J 61:767–781
Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14:1935–1940
Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PB, An G, Colombo L, Kater MM (2007) The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J 52:690–699
Dreni L, Pilatone A, Yun D, Erreni S, Pajoro A, Caporali E, Zhang D, Kater MM (2011) Functional analysis of All AGAMOUS Subfamily Members in Rice Reveals their roles in reproductive Organ identity determination and meristem determinacy. Plant Cell 23:2850–2863
Duan Y, Diao Z, Liu H, Cai M, Wang F, Lan T, Wu W (2010) Molecular cloning and functional characterization of OsJAG gene based on a complete-deletion mutant in rice (Oryza sativa L.). Plant Mol Biol 74:605–615
Ferrario S, Immink RG, Shchennikova A, Busscher-Lange J, Angenent GC (2003) The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 15:914–925
Fornara F, Parenicová L, Falasca G, Pelucchi N, Masiero S, Ciannamea S, Lopez-Dee Z, Altamura MM, Colombo L, Kater MM (2004) Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol 135:2207–2219
Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M (2002) The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett 514:351–354
Jang S, Lee B, Kim C, Kim SJ, Yim J, Han JJ, Lee S, Kim SR, An G (2003) The OsFOR1 gene encodes a polygalacturonase- inhibiting protein (PGIP) that regulates floral organ number in rice. Plant Mol Biol 53:357–372
Jeon JS, Jang S, Lee S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG, An G (2000a) Leafy hull sterile1 Is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12:871–884
Jeon JS, Lee S, Jung KH, Yang WS, Yi GH, Oh BG, An G (2000b) Production of transgenic rice plants showing reduced heading date and plant height by ectopic expression of rice MADS-box genes. Mol Breed 6:581–592
Jiang L, Qian Q, Mao L, Zhou Q, Zhai W (2005) Characterization of the Rice floral Organ number mutant fon3. J Integr Plant Biol 47:100–106
Kazama Y, Fujiwara MT, Koizumi A, Nishihara K, Nishiyama R, Kifune E, Abe T, Kawano S (2009) A SUPERMAN-like gene is exclusively expressed in female flowers of the dioecious plant Silene latifolia. Plant Cell Physiol 50:1127–1141
Khanday I, Yadav SR, Vijayraghavan U (2013) Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways. Plant Physiol 161:1970–1983
Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, Yoshida H, Nagamura Y, Kyozuka J (2012) Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24:1848–1859
Krizek BA, Lewis MW, Fletcher JC (2006) RABBIT EARS is a second-whorl repressor of AGAMOUS that maintains spatial boundaries in Arabidopsis flowers. Plant J 45:369–383
Lamb RS, Irish VF (2003) Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc Natl Acad Sci U S A 100:6558–6563
Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, Franke R, Zhang P, Chen L, Gao Y, Liang W, Zhang D (2010) Cytochrome P450 family member CYP704B2 catalyzes the ω -Hydroxylation of fatty acids and is required for anther Cutin biosynthesis and pollen Exine Formation in Rice. Plant Cell 22:173–190
Li HF, Liang WQ, Hu Y, Zhu L, Yin C, Xu J, Dreni L, Kater MM, Zhang D (2011) Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23:2536–2552
Lohmann JU, Weigel D (2002) Building beauty: the genetic control of floral patterning. Dev Cell 2:135–142
Lu S, Wei H, Wang Y, Wang H, Yang R, Zhang X, Tu J (2012) Overexpression of a transcription factor OsMADS15 modifies plant architecture and flowering time in rice (Oryza sativa L.). Plant Mol Biol Report 30:1461–1469
Mandel MA, Yanofsky MF (1995) The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell 7:1763–1771
Moon S, Jung KH, Lee DE, Lee DY, Lee J, An K, Kang HG, An G (2006) The rice FON1 gene controls vegetative and reproductive development by regulating shoot apical meristem size. Mol Cell 21:147–152
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Nagasawa N, MiyOshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y (2003) SUPERWOMAN1 and DROOPING LEAF Genes control floral organ identity in rice. Development 130:705–718
Nandi AK, Kushalappa K, Prasad K, Vijayraghavan U (2000) A conserved function for Arabidopsis SUPERMAN in regulating floral-whorl cell proliferation in rice, a monocotyledonous plant. Curr Biol 10:215–222
Nibau C, Di Stilio VS, Wu HM, Cheung AY (2011) Arabidopsis and Tobacco superman regulate hormone signalling and mediate cell proliferation and differentiation. J Exp Bot 62:949–961
Prasad K, Parameswaran S, Vijayraghavan U (2005) OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J 43:915–928
Rudall PJ, Bateman RM (2004) Evolution of zygomorphy in monocot flowers: iterative patterns and developmental constraints. New Phytol 162:25–44
Sakai H, Medrano LJ, Meyerowitz EM (1995) Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378:199–203
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108
Sentoku N, Kato H, Kitano H, Imai R (2005) OsMADS22, an STMADS11-like MADS-box gene of rice, is expressed in non-vegetative tissues and its ectopic expression induces spikelet meristem indeterminacy. Mol Gen Genomics 273:1–9
Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z (2004) Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci U S A 101:11494–11499
Sun LP, Zhang YX, Zhang PP, Zheng-fu Y, Xiao-deng Z, Xi-hong S, Zhen-hua Z, Xia H, Dan-dan X, Wei-xun W, Zi-he L, Li-yong C, Shi-hua C (2015) K-Domain splicing factor OsMADS1 regulates open hull male sterility in rice. Rice Sci 22:207–216
Suzaki T, Sato M, Ashikari M, Miyoshi M, Nagato Y, Hirano HY (2004) The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131:5649–5657
Suzaki T, Toriba T, Fujimoto M, Tsutsumi N, Kitano H, Hirano HY (2006) Conservation and diversification of meristem maintenance mechanism in Oryza sativa: function of the FLORALORGAN NUMBER2 gene. Plant Cell Physiol 47:1591–1602
Wang K, Tang D, Hong L, Xu W, Huang J, Li M, Gu M, Xue Y, Cheng Z (2010) DEP and AFO regulate reproductive habit in rice. PLoS Genet 6, e1000818
Wang H, Zhang L, Cai Q, Hu Y, Jin Z, Zhao X, Fan W, Huang Q, Luo Z, Chen M, Zhang D (2015) OsMADS32 interacts with PI-like proteins and regulates rice flower development. J Integr Plant Biol 57:504–513
Xiao H, Wang Y, Liu D, Wang W, Li X, Zhao X, Xu J, Zhai W, Zhu L (2003) Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference. Plant Mol Biol 52:957–966
Xiao H, Tang JF, Li YF, Wang W, Li X, Jin L, Xie R, Luo H, Zhao X, Meng Z (2009) STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice. Plant J 59:789–801
Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY (2004) The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16:500–509
Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, Hirano HY (2006) Functional diversification of the two C-class MADS box genes OsMADS3 and OsMADS58 in Oryza sativa. Plant Cell 18:15–18
Yuan Z, Gao S, Xue DW, Luo D, Li LT, Ding SY, Yao X, Wilson ZA, Qian Q, Zhang DB (2009) RETARDED PALEA1 controls palea development and floral zygomorphy in rice controls palea development and floral zygomorphy in rice. Plant Physiol 149:235–244
Zhang D, Wilson ZA (2009) Stamen specification and anther development in rice. Chin Sci Bull 54:2342–2353
Zhang Q, Shen BZ, Dai XK, Mei MH, Saghai Maroof MA, Li ZB (1994) Using bulked extremes and recessive class to map genes for photoperiod-sensitive genic male sterility in rice. Proc Natl Acad Sci U S A 91:8675–8679
Zheng M, Wang Y, Wang Y, Wang C, Ren Y, Lv J, Peng C, Wu T, Liu K, Zhao S, Liu X, Guo X, Jiang L, Terzaghi W, Wan J (2015) DEFORMED FLORAL ORGAN1 (DFO1) regulates floral organ identity by epigenetically repressing the expression of OsMADS58 in rice (Oryza sativa). New Phytol 206:1476–1490
Acknowledgments
We thank Professor Dabing Zhang and Wanqi Liang (Shanghai Jiao Tong University) for their kind guidance in the analysis of semi-thin sections. This work was supported by the National Special Fund for Agro-scientific Research in the Public Interest (Grants 20140302 and 20150308), the National Key Transform Program (Grants 2014ZX08001-002), the National Natural Science Foundation of China (Grants 31501290), the Zhejiang Provincial Natural Science Foundation of China (Grants Q13C130009), and the Super Rice Breeding Innovation Team and Rice Heterosis Mechanism Research Innovation Team of the Chinese Academy of Agricultural Sciences Innovation Project (Grants CAAS-ASTIP-2013-CNRRI).
Author Contributions
Shihua Cheng, Liyong Cao and Yingxin Zhang conceived and designed the study. Lianping Sun conducted the experiments andreceived help from Peipei Zhang, Zhengfu Yang, Xiaoxiao Zhou, Dandan Xuan and Zihe Li in the sampling and cytological observations. Weixun Wu, Xiaodeng Zhan, Xihong Shen, Ping Yu and Daibo Chen guided and participated in the anthesis habit analysis. Lianping Sun and Peipei Zhang collected and analyzed the data. Lianping Sun, Yingxin Zhang and Peipei Zhang wrote the paper. Lianping Sun, Yingxin Zhang, Peipei Zhang, Zhengfu Yang, Xiaoxiao Zhou, Dandan Xuan, Zihe Li, Weixun Wu, Xiaodeng Zhan, Xihong Shen, Ping Yu, Daibo Chen, Li-yong Cao and Shi-hua Cheng reviewed and edited the manuscript. All authors read and approved the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no conflict of interests regarding the publication of this paper.
Additional information
Lianping P. Sun, Yingxin X. Zhang and Peipei P. Zhang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Sun, L.P., Zhang, Y.X., Zhang, P.P. et al. Morphogenesis and Gene Mapping of deformed interior floral organ 1 (difo1), a Novel Mutant Associated with Floral Organ Development in Rice. Plant Mol Biol Rep 35, 130–144 (2017). https://doi.org/10.1007/s11105-016-1007-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-016-1007-x