Abstract
Cyanobacteria are oxygen-evolving photosynthetic prokaryotes that affect the global carbon and nitrogen turnover. Synechocystis sp. PCC 6803 (Synechocystis 6803) is a model cyanobacterium that has been widely studied and can utilize and uptake various nitrogen sources and amino acids from the outer environment and media. l-arginine is a nitrogen-rich amino acid used as a nitrogen reservoir in Synechocystis 6803, and its biosynthesis is strictly regulated by feedback inhibition. Argininosuccinate synthetase (ArgG; EC 6.3.4.5) is the rate-limiting enzyme in arginine biosynthesis and catalyzes the condensation of citrulline and aspartate using ATP to produce argininosuccinate, which is converted to l-arginine and fumarate through argininosuccinate lyase (ArgH). We performed a biochemical analysis of Synechocystis 6803 ArgG (SyArgG) and obtained a Synechocystis 6803 mutant overexpressing SyArgG and ArgH of Synechocystis 6803 (SyArgH). The specific activity of SyArgG was lower than that of other arginine biosynthesis enzymes and SyArgG was inhibited by arginine, especially among amino acids and organic acids. Both arginine biosynthesis enzyme-overexpressing strains grew faster than the wild-type Synechocystis 6803. Based on previous reports and our results, we suggest that SyArgG is the rate-limiting enzyme in the arginine biosynthesis pathway in cyanobacteria and that arginine biosynthesis enzymes are similarly regulated by arginine in this cyanobacterium. Our results contribute to elucidating the regulation of arginine biosynthesis during nitrogen metabolism.
Key message
This study revealed the catalytic efficiency and inhibition of cyanobacterial argininosuccinate synthetase by arginine and demonstrated that a strain overexpressing this enzyme grew faster than the wild-type strain.
Similar content being viewed by others
Introduction
Cyanobacteria are prokaryotes that can perform oxygen-evolving photosynthesis, and some cyanobacteria are responsible for 25% of the net oceanic productivity (Rae et al. 2013; Flombaum et al. 2013). Cyanobacteria are observed in various habitats, including freshwater, seawater, and terrestrial habitats, and are major primary producers (Hess 2011). Cyanobacteria have a variety of morphologies, being unicellular or filamentous with or without heterocysts, which are cells specialized in nitrogen fixation (Schirrmeister et al. 2013; Esteves-Ferreira et al. 2017). Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803) is one of the most studied non-nitrogen-fixing unicellular cyanobacteria (Yu et al. 2013). Synechocystis 6803 has properties of a model cyanobacterium, such as entire genome sequence information, natural transformation capability, and resistance to cryopreservation (Yu et al. 2013; Kaneko et al. 1996).
Synechocystis 6803 uses various nitrogen sources such as nitrate (NO3−), nitrite (NO2−), ammonium (NH4+), urea [CO(NH2)2], and amino acids (arginine, glutamate, and glutamine), preferentially utilizing ammonium (Quintero et al. 2001; Valladares et al. 2002; Flores et al. 2005; Muro-Pastor et al. 2005). Synechocystis 6803 grows more rapidly in the presence of ammonium than nitrate (Inabe et al. 2021). After uptake, these inorganic nitrogen sources, except for amino acids, are converted to ammonium and utilized for nitrogen assimilation via the glutamine synthetase-glutamate synthase (GS-GOGAT) cycle (Esteves-Ferreira et al. 2018; Mills et al. 2020). Glutamate is primarily consumed as a nitrogen source and is distributed in various cellular building blocks (Forchhammer and Selim 2020; Inabe et al. 2021).
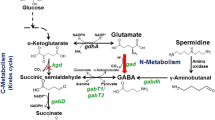
l-Arginine is synthesized from glutamate through eight enzymatic reactions similar to those in Escherichia coli (Fig. 1). Synechocystis 6803 has a bifunctional enzyme that catalyzes the first and fifth steps instead of having separate enzymes that catalyze the first and fifth steps (Mills et al. 2020). In cyanobacteria, N-acetyl-glutamate kinase (NAGK) is a second-step enzyme that is primarily subject to feedback inhibition by arginine during its biosynthesis (Hoare and Hoare 1966). Cyanobacterial argininosuccinate lyase (ArgH), the last step of arginine biosynthesis, is also inhibited by arginine, and this inhibition for cyanobacterial ArgH changes with pH (Katayama and Osanai 2022). Arginine biosynthesis is also regulated by PII proteins that are widely distributed in bacteria, eukaryotic algae, and plants, and are involved in the regulation of nitrogen assimilation and metabolism (Forchhammer 2004). Arginine accumulates 15-fold compared to the wild-type strain when a mutated PII is introduced strain in Synechocystis 6803 (Watzer et al. 2015). Arginine is used as ammonia and glutamate in the catabolizing pathway, and is a precursor for the synthesis of polyamines and cyanobacterial cyanophycin, which is a nitrogen reservoir composed of arginine and aspartate (Flores et al. 2019; Bolay et al. 2021).
Summary of arginine biosynthesis pathway around reactions catalyzed by ArgG in Synechocystis 6803. This reaction in arginine biosynthesis is based on the previous study (Flores et al. 2019). The gray ellipse represented arginine biosynthesis enzymes. The orange ellipse represented ArgG in this study detailed biochemical analysis was performed. Glu, glutamate; N-AcGlu, N-acetyl-glutamate; N-AcGluP, N-acetyl-glutamyl phosphate; N-AcGluSS, N-acetyl-glutamate semialdehyde; N-AcOrn, N-acetyl-ornithine; Orn, ornithine; Carbamoyl-P, carbamoyl phosphate; Cit, citrulline; Asp, aspartate; AS, argininosuccinate; Arg, arginine; Fum, fumarate
Argininosuccinate synthetase (ArgG; EC 6.3.4.5) catalyzes the condensation of citrulline and aspartate into argininosuccinate, the immediate precursor of arginine (Haines et al. 2011). ArgG is considered a rate-limiting step in the urea cycle and nitric oxide (NO) production in mammals (Haines et al. 2011; Jackson et al. 1986; Meijer et al. 1990). The crystal structures of ArgG have been revealed in E. coli, Thermus thermophilus, and human (Lemke and Howell 2001; Goto et al. 2002, 2003; Karlberg et al. 2008). The kinetic properties of ArgG have been reported mainly in mammals and eukaryotes such as human lymphoblasts, rat liver, and Saccharomyces cerevisiae (Kimball and Jacoby 1980; Saheki et al. 1977; Hilger et al. 1979). In Synechocystis 6803, the gene encoding argininosuccinate synthetase has been annotated as argG (Kaneko et al. 1996). A previous report has suggested that NrrA, a nitrogen-modulated response regulator, regulates ArgG levels for arginine synthesis under nitrogen excess in Synechocystis 6803 (Liu and Yang 2014). Argininosuccinate synthetase of Synechocystis 6803 from crude cell extracts shows specific activities approximately 3-4-fold lower in the NrrA deletion strain than in the wild-type strain. (Liu and Yang 2014). However, the biochemical properties of Synechocystis 6803 ArgG (SyArgG), such as the regulation by metabolites and catalytic efficiency, have not been reported in detail.
In the present study, we revealed the biochemical characteristics of SyArgG, which lowered the activity of arginine biosynthetic enzymes and was inhibited by arginine. Physiological essays revealed that Synechocystis 6803 accumulated intracellular aspartate when arginine was the sole nitrogen source.
Material & methods
Vector construction and expression of recombinant proteins
The genomic region containing the argininosuccinate synthetase open reading frame (ORF) in Synechocystis sp. PCC 6803 (slr0585) was obtained from the KEGG database (https://www.genome.jp/kegg/). A BamHI-XhoI fragment was artificially synthesized by Eurofin Genomics Co. Ltd., and the resultant fragment was cloned into the BamHI-XhoI sites of pGEX-6P-1 (GE Healthcare Japan, Tokyo, Japan).
The vector was transformed into E. coli BL21(DE3) cells (BioDynamics Laboratory Inc., Tokyo, Japan). Five L of transformed E. coli was cultivated in LB medium at 30 °C with shaking (150 rpm), and protein expression was induced overnight in the presence of 0.1 mM isopropyl-β-d-1-thiogalactopyranoside (FUJIFILM Wako Pure Chemical, Osaka, Japan).
Affinity purification of recombinant proteins
Centrifuged E. coli BL21(DE3) cells resuspended in 40 mL PBST (0.137 M NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4·12H2O, 1.47 mM KH2PO4, and 0.001% Tween 20) were disrupted by sonication (VC-750, EYELA, Tokyo, Japan) 10 times for 20 s at 20% intensity. The insoluble fraction was removed from the disrupted crude extract by centrifugation at 5800 × g for 2 min at 4 °C. The supernatant was transferred to a 50 mL tube and washed with 700 μL of Glutathione Sepharose 4B resin (Global Life Sciences Technologies Japan K.K., Tokyo, Japan), and the mixture was gently shaken for 30 min. After centrifugation, the supernatant was removed, and the resin was resuspended in 900 μL of ice-cold PBST. After washing 10 times, the recombinant proteins were eluted five times with 700 μL of GST elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM reduced glutathione). Proteins were concentrated using a Vivaspin 500 MWCO 50,000 device (Sartorius, Germany) and protein concentrations were analyzed using a Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL, USA). SDS-PAGE was performed to evaluate protein purification by staining with Quick Blue Staining Solution (BioDynamics Laboratory Inc., Tokyo, Japan).
Enzymatic assay for SyArgG
SyArgG activity was determined using 250 pmol of enzyme mixed in 1 mL of assay solution [100 mM Tris-HCl (pH 9.0), 2 mM KCl, 5 mM MgCl2, 7.5 mM citrulline, 7.5 mM sodium aspartate, 16 mM potassium phosphoenolpyruvate, 1 mM ATP, 0.2 mM NADH, 10 U myokinase, 10 U pyruvate kinase, and 10 U lactate dehydrogenase (Oriental Yeast Co., Ltd., Tokyo, Japan)]. SyArgG activity was determined based on the formation of AMP coupled with the oxidation of NADH by myokinase, pyruvate kinase, and lactate dehydrogenase (Schuegraf et al. 1960). Absorbance at 340 nm was monitored using a Hitachi U-3310 spectrophotometer (Hitachi High-Tech, Tokyo, Japan). One unit of SyArgG activity was defined as the consumption of 1 μmol of NADH per minute. Km and Vmax were calculated by curve fitting using KaleidaGraph v4.5 software, and kcat was calculated using Vmax.
Cyanobacterial strain and culture conditions
The glucose-tolerant Synechocystis sp. PCC 6803 strain (hereafter referred to as the GT strain) is isolated by Williams in 1988 (Williams 1988). Synechocystis 6803 was cultivated in 70 mL of modified BG-11 medium, consisting of BG-110 liquid medium (Rippka 1988) supplemented with 20 mM HEPES-KOH (pH 7.8) and 5 mM NaNO3 or l-arginine. For the incubation, the cultures were bubbled with 1% (v/v) CO2 in the air and incubated at 30 °C under continuous white light (50 μmol photons m− 2 s− 1). For the cultivation of the mutant strains overexpressing argG or argH, 70 mL of modified BG-11 medium containing 0.7 μg/mL kanamycin was used. The OD730 of the cyanobacterial cultures was measured using a Shimadzu UV-2700 UV-vis spectrometer (Shimadzu, Kyoto, Japan). The OD730 was set at 0.4 when the cultivation was started.
Plasmid construction and transformation of Synechocystis 6803 cells
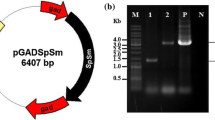
The open reading frames of Synechocystis 6803 argG (slr0585) and argH (slr1133) were synthesized and introduced into the NdeI-EcoRV sites of the pTKP2031 vector containing the psbAII promoter by Eurofin Genomics Japan. The psbAII promoter and the gene encoding SyArgG or SyArgH were integrated into the open reading frames slr2030 and slr2031 in Synechocystis 6803 genome, along with a kanamycin resistance cassette (Fig. 2a). Transformation of the GT strain was performed by homologous recombination, as described previously (Osanai et al. 2011). The generated strains overexpressing argG or argH were designated as ArgGOX and ArgHOX, respectively.
Construction of the ArgGOX and ArgHOX strains. (a) Vectors for establishing the ArgGOX and ArgHOX strains. (b) The expression levels of SyArgG and SyArgH in the GT, ArgGOX, and ArgHOX strains. The data represent the relative amounts of transcript products, and the amount in the GT strain was set at 100%. Data represent the means ± SD obtained from four or five independent experiments. (c) The expression levels of arginine biosynthesis-related genes in the GT, ArgGOX, and ArgHOX strains. The data represent the relative amounts of transcript products, and the amount in the GT strain was set at 100%. Data represent the means ± SD obtained from six independent experiments. Statistically significant differences between the activity in the absence or presence of the effector were examined by two-tailed Student’s t-test and are represented by asterisks (* = P < 0.05, ** = P < 0.005)
RNA extraction and quantitative real-time PCR
The cyanobacterial cells were harvested for 2 days of photoautotrophic cultivation [OD730 × culture volume (mL) = 15] and stored at -30 °C. Frozen cells were suspended in 1 mL ISOGEN (Nippon gene, Tokyo, Japan), gently shaken for 30 min, and then 200 μL of chloroform was added. The mixture was then strongly shaken and centrifuged at 17,900 × g, 4 °C, 15 min. Thereafter, 400 μL of isopropanol was added to 400 μL of supernatant and left on ice for 15 min. After centrifugation at 17,900 × g for 25 min at 4 °C, 200 μL of ice-cold 70% ethanol was added to the pellet to remove the supernatant. After centrifugation at 17,900 × g for 3 min at 4 °C to remove the supernatant, the residue was dried using a centrifugal evaporator CVE-2000 (EYELA, Tokyo, Japan). The pellet containing 20 μg RNA was resuspended and treated with TURBO DNase (Thermo Fisher Scientific, Rockford, IL, USA) for 1 h at 37 °C to remove residual DNA. After the enzymatic reaction, 500 μL of stabilized water and 600 μL of phenol/chloroform/isoamyl alcohol (25:24:1) were added to the solution. The solution was vigorously shaken and centrifuged at 17,900 × g for 5 min at 4 °C. Then, 550 μL of supernatant was transferred to another tube, and 55 μL of 3 M sodium acetate and 550 μL of isopropanol were added to the supernatant. The solution was then gently shaken, left for 15 min on ice, and centrifuged at 17,900 × g for 25 min at 4 °C. The supernatant was removed and 200 μL of ice-cold 70% ethanol was added to the pellet. After centrifugation at 17,900 × g for 3 min at 4 °C to remove the supernatant, the pellet was dried using a centrifugal evaporator CVE-2000. The cDNAs were synthesized using the SuperScriptIII First-Strand Synthesis System (Thermo Fisher Scientific, Rockford IL, USA) from 2 μg of extracted RNA. Quantitative real-time PCR was performed using StepOne Plus (Life Technologies, Carlsbad, CA), and the internal standard was the expression levels of rnpB encoding RNase P subunit B, as described previously (Schlebusch and Forchhammer 2010). The base sequence of the using primers for cDNAs amplification are as follows Supplemental Table 1 (Table S1).
Measurement of the SyArgG activity from cell extracts
The measurement of enzymatic activity from cell extracts was performed consistent with the previously described method (Ito et al. 2023) and added some modifications. The Synechocystis 6803 GT and ArgGOX strains cells were harvested after 3 days of photoautotrophic cultivation (50 μmol photons m− 2 s− 1) [OD730 × culture volume (ml) = 100] with 5 mM NaNO3. These cells were resuspended PBS-T and were disrupted by sonication (10 s at 20% intensity) repeated 5 times. After centrifugation (21,400×g for 5 min at 4 °C), 900 μL of supernatant was collected, and 40, 200, and 400 μg of total proteins in the supernatant were used for SyArgG activity assay. The enzymatic reaction catalyzed by SyArgG was performed in 1 mL of assay solution [100 mM Tris-HCl (pH 7.5 or 9.0), 2 mM KCl, 5 mM MgCl2, 7.5 mM citrulline, 7.5 mM sodium aspartate, 16 mM potassium phosphoenolpyruvate, 1 mM ATP, 0.2 mM NADH, 10 U myokinase, 10 U pyruvate kinase, and 10 U lactate dehydrogenase (Oriental Yeast Co., Ltd., Tokyo, Japan)]. The assay solution was incubated at 38 °C for 5 min without sodium aspartate, added to the solution sodium aspartate. SyArgG activity was determined based on the formation of AMP coupled with the oxidation of NADH by myokinase, pyruvate kinase, and lactate dehydrogenase (Schuegraf et al. 1960). Absorbance at 340 nm was monitored using a Hitachi U-3310 spectrophotometer (Hitachi High-Tech, Tokyo, Japan). As a control, we measured the change of absorbance at 340 nm in the absence of sodium aspartate.
Extraction and measurement of aspartate and glutamate from Synechocystis 6803 cells
After 3 days of cultivation, Synechocystis 6803 cells [OD730 × culture volume (mL) = 100] were harvested by centrifugation [5800 × g for 4 min at 25 °C]. The cells were suspended in 600 μL of 60% (v/v) methanol and mixed with TWIN MIXER TM-282 (Asone, Osaka, Japan) for 15 min. The suspension was then centrifuged. A volume of 500 μL of the supernatant was transferred to an Amicon Ultra 3-kDa-cutoff filter (Merck, Billerica, MA, USA) and centrifuged. After centrifugation, 350 μL of the filtered fraction was dried using a CVE-2000 centrifugal evaporator (EYELA, Tokyo, Japan). The pellet was dissolved in 200 μL of 100 mM Tris-HCl buffer (pH 8.0). The quantification of aspartate and glutamate was performed using the Aspartate Assay Kit (MAK095) (Sigma-Aldrich, St. Louis, MO, USA) and F-kit l-glutamate (J.K. International, Tokyo, Japan).
Extraction cyanophycin and arginine quantification
Cyanophycin contains equal molecules of arginine and aspartate, therefore the concentration of cyanophycin equals the amount of arginine. We performed cyanophycin extraction and arginine quantification using the Sakaguchi reaction according to the studies (Burgstaller et al. 2022, Messineo 1966) with some modifications. After 3 days of cultivation, Synechocystis 6803 cells [OD730 × culture volume (mL) = 25] were harvested by centrifugation [5800 × g for 4 min at 25 °C] and 1 mL acetone was added to the cells and incubation with shaking for 30 min. After centrifugation [13,000 × g for 10 min at 25 °C], the supernatant was removed and the residue was resuspended in 1.2 mL of 0.1 M HCl. The samples were incubated and shaken for 60 min at 60. After centrifugation [13,000 × g for 10 min at 4 °C], the supernatant was transferred to another 1.5 mL microtube, and added 300 μL of 1 M Tris-HCl (pH9.0) to the supernatant. The sample was incubated on ice for 40 min. After centrifugation [18,000 × g for 15 min at 4 °C], the supernatant was removed and resuspended in 1 mL of 0.01 M HCl. The solution was used for the arginine quantification.
Arginine standards with concentrations of 0.06, 0.3 0.6, 1.2, 2.4, 4.8, 6.0 mM in 0.01 M HCl were prepared. 166 μL of reagent A [300 mg potassium iodide in 100 mL distilled water] was added to 166 μL the sample or standard. Subsequently, 500 μL of reagent B [100 mL of 5 M KOH, 2 g potassium sodium tartrate, 0.1 g 2,4-dichloro-1-naphthol, 180 mL absolute ethanol, 0.2 mL NaClO] was added to the sample and was incubated for 60 min at laboratory temperature. After incubation, 166 μL of reagent C [20% (v/v) NaClO with distilled water] was added to the sample and was incubated for 10 min. We measured the absorbance at 520 nm of the sample, using the sample without arginine as a blank.
Statistical analysis
P-values were calculated using a paired two-tailed Student’s t-test in Microsoft Excel for Windows (Redmond, WA, USA). All results were obtained from multiple independent experiments (described in detail in individual figure legends).
Results
Determination of the kinetics parameters of SyArgG and its strong inhibition by arginine
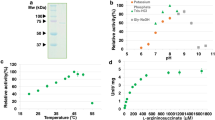
To evaluate the biochemical properties of SyArgG, glutathione-S-transferase-tagged (GST-tagged) SyArgG was expressed in E. coli cells, and GST-tagged SyArgG was purified from the soluble fraction using affinity chromatography. Purified GST-tagged SyArgG was confirmed as a single protein band using 12% SDS-PAGE (Fig. 3a). First, we examined the temperature dependence of SyArgG. The enzyme showed the highest enzymatic activity at 35–40 °C and inactivated above 50 °C (Fig. 3b). We then examined the pH range of SyArgG activity using various buffers, MES-NaOH, Tris-HCl, CHES-KOH, and CAPS-KOH (Fig. 3c).
Purification of SyArgG and determination of its optimal temperature and pH. (a) Purified GST-tagged SyArgG separated on 12% SDS-PAGE gel and stained with Quick Staining Solution. (b) Effect of temperature on SyArgG activity. (c) Effect of pH on SyArgG activity in various buffers. Blue circles, orange squares, gray triangles, and yellow rhombuses represent MES-NaOH, Tris-HCl, CHES-KOH, and CAPS-KOH, respectively
Kinetic analyses of SyArgG based on its saturation curves under 38 °C and pH 9.0 (Fig. 3b and c) for citrulline, aspartate, and ATP, respectively were subsequently performed (Fig. 4a and c). The Km, kcat, and kcat/Km values of SyArgG for citrulline were 0.35 mM, 0.69 s− 1, and 1.97 s− 1 mM− 1, respectively (Table 1). The Km, kcat, and kcat/Km values of SyArgG for aspartate were 0.36 mM, 0.77 s− 1, and 2.16 s− 1 mM− 1, respectively (Table 1). The Km, kcat, and kcat/Km values of SyArgG for ATP were 0.26 mM, 0.7 s− 1, and 2.75 s− 1 mM− 1, respectively (Table 1).
Determination of Km values for aspartate, citrulline, and ATP. and the effect of various metabolites on SyArgG activity. This measurement was performed at 38 °C and pH 9.0. SyArgG activity was measured by varying (a) sodium aspartate, (b) citrulline, and (c) ATP concentrations. One unit of SyArgG activity was defined as a consumption of 1 μmol NADH per minute. Data represent the means ± SD obtained from the triplicated independent experiments
We measured SyArgG activity in the presence of several metabolites to determine their effects. SyArgG activity was measured in the presence of selected organic and amino acids that are closely related to the tricarboxylic acid cycle and arginine biosynthesis (Fig. 5). SyArgG activity decreased to 53.9% and 18.4% in the presence of 1 mM and 5 mM arginine, respectively (Fig. 5). SyArgG activity decreased to 80.1, 78.1, and 92.1% in the presence of 5 mM ornithine, lysin, and succinate, respectively (Fig. 5). However, SyArgG activity did not decrease in the presence of glutamate, glutamine, or asparagine, which have structures similar to that of aspartate (Fig. 5).
Effect of various metabolites on SyArgG activity. This measurement was performed at 38 °C and pH 9.0 and fixed concentrations of aspartate, citrulline, and ATP were 7.5, 7.5, and 1.0 mM respectively. SyArgG activity was represented by relative activity, whereby the activity in the absence of metabolites was set at 100%. Data represent the means ± SD obtained from triplicate independent experiments. Statistically significant differences between the activity in the absence or presence of the effector were examined by two-tailed Student’s t-test and are represented by asterisks (* = P < 0.05, ** = P < 0.005, *** = P < 0.0005). Glu, glutamate; Gln, glutamine; Asn, asparagine; Orn, ornithine; Arg, arginine; Lys, lysin; Pro, proline; 2-OG, 2-oxoglutarate; Cit, citrate; Mal, malate; Fum, fumarate; Suc, succinate; Isoc, isocitrate
Synechocystis 6803 cells accumulate aspartate in the medium with arginine
To reveal the effects of arginine biosynthesis gene overexpression and different nitrogen sources on growth, we evaluated the GT strains overexpressing the SyArgG (slr0585) and SyArgH (slr1133) genes (designated ArgGOX and ArgHOX) using 5 mM NaNO3 as a nitrogen source (Fig. 2a). The RNA expression of the gene encoding SyArgG in the ArgGOX strain was 2.0-fold higher than that in the GT strain (Fig. 2b). We tried to measure the SyArgG activity from cell extracts of GT and ArgGOX strains, but the enzymatic activity was not detected, because of low activity of SyArgG. The RNA expression level of the gene encoding SyArgH in the ArgHOX strain was 40.8-fold higher than that in the GT strain (Fig. 2b). And we measured RNA expression of the arginine biosynthesis-related genes, such as the nrtA encoding nitrate transporter subunit, the aspC encoding aspartate aminotransferase, the argJ encoding the bifunctional enzyme of arginine biosynthesis, the argD encoding the bifunctional aminotransferase catalyzed arginine biosynthesis and GABA metabolism. The expression levels of the gene encoding argJ (sll1883) and nrtA (sll1450) in the ArgGOX strain were 0.53- and 0.32-fold lower than those in the GT strain, respectively (Fig. 2c). The expression levels of the gene encoding argD (slr1022) and aspC (sll0402) were not significantly different among the GT, ArgGOX, and ArgHOX strains (Fig. 2c). We also measured RNA expression of the argB gene encoding NAGK that is a significant regulator subjected to arginine feedback inhibition. The expression level of the argB encoding NAGK in the ArgGOX strain was 0.37-fold lower than in the GT strain (Fig. S1). The growth of the ArgGOX and ArgHOX strains was faster than that of the GT strain under photoautotrophic conditions with 5 mM NaNO3 as a nitrogen source (Fig. 6a). The cell densities of ArgGOX and ArgHOX strains at 72 h were 34% and 24% increase compared to GT strain, respectively (Fig. 6a). In contrast, the growth of these strains was similar under photoautotrophic conditions with 5 mM arginine as a nitrogen source (Fig. 6b). The RNA expression level of the gene encoding SyArgG in the ArgGOX strain and the gene encoding SyArgH in the ArgHOX strain was 2.1-fold and 49.6-fold higher than those in the GT strain using as a 5 mM arginine (Fig. S1). The RNA expression levels of other arginine biosynthesis-related genes showed no significant change between the GT strain and ArgGOX or ArgHOX strains (Fig. S2).
Growth curves of the GT, ArgGOX, and ArgHOX strains under photoautotrophic conditions with (a) NaNO3 and (b) arginine as nitrogen source and intracellular aspartate concentration. The blue circle, orange square, and gray triangle represent the GT, ArgGOX, and ArgHOX strains, respectively. (c) Intracellular aspartate concentration in the GT, ArgGOX, and ArgHOX strains with different nitrogen sources. Data represent the means ± SD obtained from triplicate independent experiments. Statistically significant differences between the OD730 in the GT, ArgGOX, or ArgHOX strains were examined by paired two-tailed Student’s t-tests and are represent by asterisks (* = P < 0.05, ** = P < 0.005)
Intracellular aspartate levels in the GT, ArgGOX, and ArgHOX strains under photoautotrophic conditions with 5 mM NaNO3 or arginine as the nitrogen source was then determined (Fig. 6c). The intracellular aspartate concentration with 5 mM NaNO3 was 0.06, 0.05, and 0.06 μM/OD in the GT, ArgGOX, and ArgHOX strains, respectively (Fig. 6c). The intracellular aspartate concentration with 5 mM arginine were 0.30, 0.26, and 0.24 μM/OD in the GT, ArgGOX, and ArgHOX strains, respectively (Fig. 6c). The aspartate concentration in each strain with 5 mM arginine was 5.21, 5.18, and 4.21-fold higher than that in the strains with 5 mM NaNO3 (Fig. 6c). We determined intracellular arginine from purified cyanophycin under photoautotrophic conditions with 5 mM NaNO3 or arginine as nitrogen source. The intracellular arginine of GT and ArgGOX, ArgHOX strains were 0.27 and 0.26, 0.26 mM, respectively in 5 mM NaNO3 (Fig. S3). Intracellular arginine of GT and ArgGOX, ArgHOX strains were 0.80 and 0.88, 0.88 mM, respectively in 5 mM arginine (Fig. S3). We determined intracellular glutamate under photoautotrophic conditions with 5 mM NaNO3 or arginine as a nitrogen source. The intracellular glutamate of GT, ArgGOX, and ArgHOX strains were 2.56, 1.85, and 3.24 mM, respectively in 5 mM NaNO3 (Fig. S4). Intracellular glutamate of GT, ArgGOX, and ArgHOX strains were 2.44, 2.89, and 3.98 mM, respectively in 5 mM arginine (Fig. S4).
Discussion
In this study, we revealed the biochemical properties of SyArgG, a rate-limiting enzyme in the arginine biosynthesis pathway, and that the ArgGOX and ArgHOX strains had different cell densities compared to those of the GT strain.
The Km values of SyArgG for citrulline and aspartate were similar (Table 1). The Km values of argininosuccinate synthetase of the higher plant Pinus pinaster Ait. (PpASSY) are both 0.12 mM for citrulline and aspartate (Urbano-Gámez et al. 2020). In mammals, the Km values of argininosuccinate synthetase for citrulline and aspartate are 0.02 and 0,078 in rats and 0.044 and 0.068 mM in humans, respectively (Saheki et al. 1977; Kimball and Jacoby 1980). Compared with these reports, our results showed that the Km value of cyanobacterial argininosuccinate synthetase was higher than that of the eukaryotic argininosuccinate synthetase. This suggests that Synechocystis 6803 can synthesize a precursor of arginine only under conditions of sufficient nitrogen concentration because of the low substrate affinity of SyArgG. The specific activity of SyArgG towards the substrates was lower than 1 U/mg (Fig. 4). The specific activity of the key enzyme in arginine biosynthesis, NAGK, is higher than 3 U/mg in the absence of arginine in Synechocystis 6803 (Bolay et al. 2021). In addition, the activity of the arginine biosynthesis key enzyme of Synechocystis 6803, N-acetylornithine aminotransferase, for N-acetylornithine (kcat/Km: 19.3 s− 1 mM− 1) was approximately 10-fold higher than that of SyArgG (kcat/Km: 1.97 s− 1mM− 1) (Li et al. 2023; Table 1). Synechocystis 6803 cell crude extract experiments also show that N-acetylornithine aminotransferase activity is 2-fold higher than argininosuccinate synthetase activity (Liu and Yang 2014). The kcat/Km of SyArgG for the substrate (citrulline: 1.97 s− 1mM− 1, aspartate: 2.16 s− 1mM− 1) was lower than that of the last-step enzyme in arginine biosynthesis (SyArgH: 37.3 s− 1mM− 1) (Katayama and Osanai 2022). These results indicate that SyArgG is the rate-limiting enzyme in arginine biosynthesis.
SyArgG enzymatic activity was significantly inhibited by arginine in a concentration-dependent manner (Fig. 5). Argininosuccinate synthetase activities of the thermophilic eubacteria and archaeobacteria Thermus aquaticus, Thermotoga maritima, and Sulfolobus solfataricus are also inhibited by 90% by 8-, 4-, and 2-mM arginine, respectively (Van de Casteele et al. 1990). PpASSY activity is significantly inhibited by 10 mM arginine by approximately 20%, and PpASSY activity is similarly inhibited by arginine concentrations ranging from 1 mM to above 20 mM (Urbano-Gámez et al. 2020). These results indicate that SyArgG is more sensitively regulated by feedback inhibition than PpASSY. This arginine inhibition is similar to that of previously reported arginine biosynthesis enzymes (NAGK and SyArgH) in cyanobacteria (Hoare and Hoare 1966; Bolay et al. 2021; Katayama and Osanai 2022). Under physiological conditions, the intracellular arginine concentration in Synechocystis 6803 is 0.6 mM (Zhang et al. 2018). The intracellular arginine levels in Synechocystis 6803 change depending on the growth medium (Iijima et al. 2020). We propose that arginine biosynthesis in Synechocystis 6803 is regulated by SyArgG, including the ornithine cycle, and not only by NAGK to adapt to various environments.
Under photoautotrophic conditions, ArgGOX and ArgHOX grew faster than the GT strain with 5 mM NaNO3 as a nitrogen source (Fig. 6a). This phenotype indicates that overexpression of arginine biosynthesis genes relieves the enzymatic rate-limiting step using NaNO3 as a nitrogen source. The mutated PII protein introduced strain shows a similar cell density to the wild-type strain with nitrate as a nitrogen source (Watzer et al. 2015). The nrrA deletion mutant has a similar growth rate during the exponential phase, but a longer lag phase leading up to the exponential phase (Liu and Yang 2014). When 5 mM NaNO3 was used as the nitrogen source, direct overexpression of arginine biosynthesis genes showed an increase in cell density (Fig. 6a). The argJ, nrtA, and argB expression levels were decreased in the ArgGOX strain compared to the GT strain with 5 mM NaNO3 condition (Fig. 2 and S1). The argB encoding NAGK that enzyme reaction uses ATP, similar to nrtA encoding nitrate transporter subunit. SyArgG also used ATP for the condensation of citrulline and aspartate (Fig. 4). The argJ encoded ArgJ, the enzyme introduces glutamate into arginine biosynthesis and ornithine into the ornithine cycle. We suggested that arginine biosynthesis may regulate to balance of ATP consumption. We suggested that argG overexpression controlled the introduction of new substrates into arginine biosynthesis and reduced unnecessary ATP consumption. In contrast, the cell densities of the three strains were similar when 5 mM arginine was used as the nitrogen source (Fig. 6b). Synechocystis 6803 produces and accumulates cyanophycin, resulting in increased intracellular arginine levels in BG-11 medium supplemented with 5 mM arginine and 10 mM glucose (Burgstaller et al. 2022). In this study, the intracellular arginine of these strains with 5 mM arginine was higher than those with 5 mM NaNO3 (Fig. S3). Synechocystis 6803 can uptake arginine from the media, and its uptake activity is 31-fold higher than that of the nitrogen-fixing heterocystous cyanobacteria Nostoc (Anabaena) sp. PCC 7120 (Montesinos et al. 1997). The majority of arginine taken up from the medium is incorporated into cyanophycin (Stephan et al. 2000). These results suggest that Synechocystis 6803 uptakes arginine from the medium and arginine is incorporated into cyanophycin, and intracellular arginine inhibits SyArgG and other arginine biosynthesis enzymes, resulting in attenuation of the overexpression effects. Furthermore, intracellular aspartate accumulated in all strains when 5 mM arginine was used as the nitrogen source (Fig. 6c). But intracellular glutamate decreased in the ArgGOX strain with 5 mM NaNO3 and increased with 5 mM arginine (Fig. 4S). The transcript levels of glutamine synthetase were increased, while transcript levels of glutamine synthetase-inactivating factor IF7 (gifA) and IF17 (gifB) with arginine-grown WT strain versus nitrate-grown WT strain (Schriek et al. 2008). The ArgGOX strain with 5 mM NaNO3 occurs a similar phenomenon, consequently, the intracellular glutamate of the ArgGOX strain was decreased. The amino acid profile of Synechocystis 6803 in artificial seawater showed that the levels of ornithine and aspartate were higher than those in cells grown in BG-11 medium (Iijima et al. 2015). SyArgG activity decreased by 80% in the presence of 5 mM ornithine (Fig. 5). We suggest that arginine biosynthesis enzymes, including SyArgG, are inhibited by intracellular arginine and that the precursor aspartate is not used as a substrate for SyArgG and accumulates in Synechocystis 6803 cells. We may hypothesize arginine biosynthesis gene overexpression and using arginine as a nitrogen source affects other amino acid metabolism.
In this study, we demonstrated that SyArgG is the rate-limiting step in arginine biosynthesis in Synechocystis 6803. This study contributes to the elucidation of nitrogen metabolism, regulation of arginine biosynthesis, and not only NAGK regulation in Synechocystis 6803. Under photoautotrophic conditions, the ArgGOX and ArgHOX strains grew faster than the GT strain. Optimized nitrogen and phosphorus supply yielded a higher productivity of glycogen and PHB (poly-3-hydroxybutyrate) and had little negative effect on biomass production (Singhon et al. 2021). These arginine biosynthesis-overexpressing strains will increase biomass and polyamine productivity in Synechocystis 6803.
Data availability
Not applicable.
References
Bolay P, Rozbeh R, Muro-Pastor MI, Timm S, Hagemann M, Florencio FJ, Forchhammer K, Klähn S (2021) The novel PII-interacting protein PirA controls flux into the cyanobacterial ornithine-ammonia cycle. mBio 12(2):e00229–e00221. https://doi.org/10.1128/mbio.00229-21
Burgstaller H, Wang Y, Caliebe J, Hueren V, Appel J, Boehm M, Leitzke S, Theune M, King PW, Gutekunst K (2022) Synechocystis sp. PCC 6803 requires bidirectional hydrogenase to metabolize glucose and arginine under oxic conditions. Front Microbiol 13:896190. https://doi.org/10.3389/fmicb.2022.896190
Esteves-Ferreira AA, Cavalcanti JHF, Vaz MGMV, Alvarenga LV, Nunes-Nesi A, Araújo WL (2017) Cyanobacterial nitrogenases: phylogenetic diversity, regulation, and functional predictions. Genet Mol Biol 40:261–275. https://doi.org/10.1590/1678-4685-GMB-2016-0050
Esteves-Ferreira AA, Inaba M, Fort A, Araújo WL, Sulpice R (2018) Nitrogen metabolism in cyanobacteria: metabolic and molecular control, growth consequences and biotechnological applications. Crit Rev Microbiol 44:541–560. https://doi.org/10.1080/1040841X.2018.1446902
Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N, Karl DM, Li WK, Lomas MW, Veneziano D, Vera CS, Vrugt JA, Martiny AC (2013) Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA 110:9824–9829. https://doi.org/10.1073/pnas.1307701110
Flores E, Frías JE, Rubio LM, Herrero A (2005) Photosynthetic nitrate assimilation in cyanobacteria. Photosynth Res 83:117–133. https://doi.org/10.1007/s11120-004-5830-9
Flores E, Arévalo S, Burnat M (2019) Cyanophycin and arginine metabolism in cyanobacteria. Algal Res 42:101577. https://doi.org/10.1016/j.algal.2019.101577
Forchhammer K (2004) Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol Rev 28:319–333. https://doi.org/10.1016/j.femsre.2003.11.001
Forchhammer K, Selim KA (2020) Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol Rev 44:33–53. https://doi.org/10.1093/femsre/fuz025
Goto M, Nakajima Y, Hirotsu K (2002) Crystal structure of argininosuccinate synthetase from Thermus thermophilus HB8. Structural basis for the catalytic action. J Biol Chem 277:15890–15896. https://doi.org/10.1074/jbc.M112430200
Goto M, Omi R, Miyahara I, Sugahara M, Hirotsu K (2003) Structures of argininosuccinate synthetase in enzyme-ATP substrates and enzyme-AMP product forms: stereochemistry of the catalytic reaction. J Biol Chem 278:22964–22971. https://doi.org/10.1074/jbc.M213198200
Haines RJ, Pendleton LC, Eichler DC (2011) Argininosuccinate synthase: at the center of arginine metabolism. Int J Biochem Mol Biol 2:8–23
Hess WR (2011) Cyanobacterial genomics for ecology and biotechnology. Curr Opin Microbiol 14:608–614. https://doi.org/10.1016/j.mib.2011.07.024
Hilger F, Simon JP, Stalon V (1979) Yeast argininosuccinate synthetase. Purification; structural and kinetic properties. Eur J Biochem 94:153–163. https://doi.org/10.1111/j.1432-1033.1979.tb12882.x
Hoare DS, Hoare SL (1966) Feedback regulation of arginine biosynthesis in blue-green algae and photosynthetic bacteria. J Bacteriol 92:375–379. https://doi.org/10.1128/jb.92.2.375-379.1966
Iijima H, Nakaya Y, Kuwahara A, Hirai MY, Osanai T (2015) Seawater cultivation of freshwater cyanobacterium Synechocystis sp. PCC 6803 drastically alters amino acid composition and glycogen metabolism. Front Microbiol 6:326. https://doi.org/10.3389/fmicb.2015.00326
Iijima H, Watanabe A, Sukigara H, Shirai T, Kondo A, Osanai T (2020) Simultaneous increases in the levels of compatible solutes by cost-effective cultivation of Synechocystis sp. PCC 6803. Biotechnol Bioeng 117:1649–1660. https://doi.org/10.1002/bit.27324
Inabe K, Miichi A, Matsuda M, Yoshida T, Kato Y, Hidese R, Kondo A, Hasunuma T (2021) Nitrogen availability affects the metabolic profile in cyanobacteria. Metabolites 11:867. https://doi.org/10.3390/metabo11120867
Ito S, Watanabe A, Osanai T (2023) Regulation of l-aspartate oxidase contributes to NADP + biosynthesis in Synechocystis sp. PCC 6803. Plant Physiol 4:kiad580. https://doi.org/10.1093/plphys/kiad580
Jackson MJ, Beaudet AL, O’Brien WE (1986) Mammalian urea cycle enzymes. Annu Rev Genet 20:431–464. https://doi.org/10.1146/annurev.ge.20.120186.002243
Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3:109–136. https://doi.org/10.1093/dnares/3.3.109
Karlberg T, Collins R, van den Berg S, Flores A, Hammarström M, Högbom M, Holmberg Schiavone L, Uppenberg J (2008) Structure of human argininosuccinate synthetase. Acta Crystallogr D Biol Crystallogr 64:279–286. https://doi.org/10.1107/S0907444907067455
Katayama N, Osanai T (2022) Arginine inhibition of the argininosuccinate lyases is conserved among three orders in cyanobacteria. Plant Mol Biol 110:13–22. https://doi.org/10.1007/s11103-022-01280-x
Kimball ME, Jacoby LB (1980) Purification and properties of argininosuccinate synthetase from normal and canavanine-resistant human lymphoblasts. Biochemistry 19:705–709. https://doi.org/10.1021/bi00545a015
Lemke CT, Howell PL (2001) The 1.6 a crystal structure of E. Coli argininosuccinate synthetase suggests a conformational change during catalysis. Structure 9:1153–1164. https://doi.org/10.1016/S0969-2126(01)00683-9
Li ZM, Bai F, Wang X, Xie C, Wan Y, Li Y, Liu J, Li Z (2023) Kinetic characterization and catalytic mechanism of N-acetylornithine aminotransferase encoded by slr1022 gene from Synechocystis sp. PCC 6803. Int J Mol Sci 24:5853. https://doi.org/10.3390/ijms24065853
Liu D, Yang C (2014) The nitrogen-regulated response regulator NrrA controls cyanophycin synthesis and glycogen catabolism in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem 289:2055–2071. https://doi.org/10.1074/jbc.M113.515270
Meijer AJ, Lamers WH, Chamuleau RA (1990) Nitrogen metabolism and ornithine cycle function. Physiol Rev 70(3):701–748. https://doi.org/10.1152/physrev.1990.70.3.701
Messineo L (1966) Modification of the Sakaguchi reaction: Spectrophotometric determination of arginine in proteins without previous hydrolysis. Arch Biochem Biophys 117:534–540.https://doi.org/10.1016/0003-9861(66)90094-4
Mills LA, McCormick AJ, Lea-Smith DJ (2020) Current knowledge and recent advances in understanding metabolism of the model cyanobacterium Synechocystis sp. PCC 6803. Biosci Rep 40:BSR20193325. https://doi.org/10.1042/BSR20193325
Montesinos ML, Herrero A, Flores E (1997) Amino acid transport in taxonomically diverse cyanobacteria and identification of two genes encoding elements of a neutral amino acid permease putatively involved in recapture of leaked hydrophobic amino acids. J Bacteriol 179:853–862. https://doi.org/10.1128/jb.179.3.853-862.1997
Muro-Pastor MI, Reyes JC, Florencio FJ (2005) Ammonium assimilation in cyanobacteria. Photosynth Res 83:135–150. https://doi.org/10.1007/s11120-004-2082-7
Osanai T, Oikawa A, Azuma M, Tanaka K, Saito K, Hirai MY, Ikeuchi M (2011) Genetic engineering of group 2 sigma factor SigE widely activates expressions of sugar catabolic genes in Synechocystis species PCC 6803. J Biol Chem 286:30962–30971. https://doi.org/10.1074/jbc.M111.231183
Quintero MJ, Montesinos ML, Herrero A, Flores E (2001) Identification of genes encoding amino acid permeases by inactivation of selected ORFs from the Synechocystis genomic sequence. Genome Res 11:2034–2040. https://doi.org/10.1101/gr.196301
Rae BD, Long BM, Whitehead LF, Förster B, Badger MR, Price GD (2013) Cyanobacterial carboxysomes: microcompartments that facilitate CO2 fixation. J Mol Microbiol Biotechnol 23:300–307. https://doi.org/10.1159/000351342
Rippka R (1988) Isolation and purification of cyanobacteria. Methods Enzymol 167:3–27. https://doi.org/10.1016/0076-6879(88)67004-2
Saheki T, Kusumi T, Takeda S, Katsunuma T (1977) Studies of rat liver argininosuccinate synthetase I. Physicochemical, catalytic, and immunochemical properties. J Biochem 81:687–696
Schirrmeister BE, de Vos JM, Antonelli A, Bagheri HC (2013) Evolution of multicellularity coincided with increased diversification of cyanobacteria and the great oxidation event. Proc Natl Acad Sci U S A 110:1791–1796. https://doi.org/10.1073/pnas.1209927110
Schlebusch M, Forchhammer K (2010) Requirement of the nitrogen starvation-induced protein Sll0783 for polyhydroxybutyrate accumulation in Synechocystis sp. strain PCC 6803. Appl Environ Microbiol 76:6101–6107. https://doi.org/10.1128/AEM.00484-10
Schriek S, Aguirre-von-Wobeser E, Nodop A, Becker A, Ibelings BW, Bok J, Staiger D, Matthijs HC, Pistorius EK, Michel KP (2008) Transcript profiling indicates that the absence of PsbO affects the coordination of C and N metabolism in Synechocystis sp. PCC 6803. Physiol Plant 133(3):525–543. https://doi.org/10.1111/j.1399-3054.2008.01119.x
Schuegraf A, Ratner S, Warner RC (1960) Free energy changes of the argininosuccinate synthetase reaction and of the hydrolysis of the inner pyrophosphate bond of adenosine triphosphate. J Biol Chem 235:3597–3602. https://doi.org/10.1016/S0021-9258(18)64515-6
Singhon P, Phoraksa O, Incharoensakdi A, Monshupanee T (2021) Increased bioproduction of glycogen, lipids, and poly(3-hydroxybutyrate) under partial supply of nitrogen and phosphorus by photoautotrophic cyanobacterium Synechocystis sp. PCC 6803. J Appl Phycol 33:2833–2843. https://doi.org/10.1007/s10811-021-02494-0
Stephan DP, Ruppel HG, Pistorius EK (2000) Interrelation between cyanophycin synthesis, L-arginine catabolism and photosynthesis in the cyanobacterium Synechocystis sp. strain PCC 6803. Z naturforsch C. J Biosci 55(11–12):927–942. https://doi.org/10.1515/znc-2000-11-1214
Urbano-Gámez JA, El-Azaz J, Ávila C, de la Torre FN, Cánovas FM (2020) Enzymes involved in the biosynthesis of arginine from ornithine in maritime pine (Pinus pinaster Ait). Plants (Basel) 9:1271. https://doi.org/10.3390/plants9101271
Valladares A, Montesinos ML, Herrero A, Flores E (2002) An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol Microbiol 43:703–715. https://doi.org/10.1046/j.1365-2958.2002.02778.x
Van de Casteele M, Desmarez M, Legrain C, Piérard A, Glansdorff N (1990) Pathways of arginine biosynthesis in extreme thermophilic archaeo- and eubacteria. J Gen Microbiol 136:1177–1183. https://doi.org/10.1099/00221287-136-7-1177
Watzer B, Engelbrecht A, Hauf W, Stahl M, Maldener I, Forchhammer K (2015) Metabolic pathway engineering using the central signal processor PII. Microb Cell Fact 14:192. https://doi.org/10.1186/s12934-015-0384-4
Williams JGK (1988) Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol 167:766–778. https://doi.org/10.1016/0076-6879(88)67088-1
Yu Y, You L, Liu D, Hollinshead W, Tang YJ, Zhang F (2013) Development of Synechocystis sp. PCC 6803 as a phototrophic cell factory. Mar Drugs 11:2894–2916. https://doi.org/10.3390/md11082894
Zhang H, Liu Y, Nie X, Liu L, Hua Q, Zhao GP, Yang C (2018) The cyanobacterial ornithine-ammonia cycle involves an arginine dihydrolase. Nat Chem Biol 14:575–581. https://doi.org/10.1038/s41589-018-0038-z
Funding
This work was supported by the following grants to NK from JSPS KAKENHI Grant-in-Aid for JSPS Fellows (grant numbers 21J20651 and 22KJ2845), grants to TO from JSPS KAKENHI Grant-in-Aid for Scientific Research (B) (grant number 20H02905), JST-ALCA of the Japan Science and Technology Agency (grant number JPMJAL1306), and the Asahi Glass Foundation.
Open Access funding provided by Meiji University.
Author information
Authors and Affiliations
Contributions
N.K. designed the study, performed the experiments, analyzed the data, and wrote the manuscript. T.O. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Katayama, N., Osanai, T. Arginine inhibits the arginine biosynthesis rate-limiting enzyme and leads to the accumulation of intracellular aspartate in Synechocystis sp. PCC 6803. Plant Mol Biol 114, 27 (2024). https://doi.org/10.1007/s11103-024-01416-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-024-01416-1