Abstract
The enzyme poly(ADP-ribose)polymerase (PARP) has a dual function being involved both in the poly(ADP-ribosyl)ation and being a constituent of the NAD+ salvage pathway. To date most studies, both in plant and non-plant systems, have focused on the signaling role of PARP in poly(ADP-ribosyl)ation rather than any role that can be ascribed to its metabolic function. In order to address this question we here used a combination of expression, transcript and protein localization studies of all three PARP isoforms of Arabidopsis alongside physiological analysis of the corresponding mutants. Our analyses indicated that whilst all isoforms of PARP were localized to the nucleus they are also present in non-nuclear locations with parp1 and parp3 also localised in the cytosol, and parp2 also present in the mitochondria. We next isolated and characterized insertional knockout mutants of all three isoforms confirming a complete knockout in the full length transcript levels of the target genes as well as a reduced total leaf NAD hydrolase activity in the two isoforms (PARP1, PARP2) that are highly expressed in leaves. Physiological evaluation of the mutant lines revealed that they displayed distinctive metabolic and root growth characteristics albeit unaltered leaf morphology under optimal growth conditions. We therefore conclude that the PARP isoforms play non-redundant non-nuclear metabolic roles and that their function is highly important in rapidly growing tissues such as the shoot apical meristem, roots and seeds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nicotinamide adenine dinucleotide (NAD+) and its derivatives play a critical role in various metabolic events for maintaining energy homeostasis in living organisms. Given the fact that the reactions utilizing NAD+ are manifold it is unsurprising that redundancy exists in its pathways of biosynthesis with different routes to the same end, namely the de novo pathway and the salvage pathway (Hashida et al. 2009; Pétriacq et al. 2012). The de novo pathway comprises of five enzyme catalyzed reactions beginning with the conversion of aspartate to quinolate via the concerted action of aspartate oxidase and quinolate synthase (Schippers et al. 2008), through quinolinate phosphoribosyltransferase and nicotinate (and/or nicotinamide) mononucleotide adenylyltransferase prior to the conversion of nicotinate adenine dinucleotide (NaAD) to NAD+ by NAD synthase (NADS) (Hashida et al. 2009). Concomitantly, the four-step salvage pathway promotes the degradation of NAD+ to nicotinamide (NaM) as a result of (cyclic)ADP-ribose generation (via ADPR-cyclase), poly(ADP-ribosyl)ation (by PARP) or alternatively during the course of protein deacetylation (by SRT2) (Hunt et al. 2004; König et al. 2014). NaM is then deamidated to nicotinic acid (Na), which is transferred onto 5′-phosphoribosyl-1-pyrophosphate (PRPP) via nicotinate phosphoribosyl-transferase (NAPRT). The resultant NaMN subsequently acts as substrate for nicotinamide mononucleotide adenylyltransferease (NaMNAT), a de novo pathway enzyme, to produce nicotinate adenine dinucleotide (NaAD), which is then recycled to NAD+ (Hunt et al. 2004), thus closing the cycle. Intriguingly, this recycling system in Arabidopsis is quite different from the well-known two-step salvage pathway in mammals, where NAD+ is a product of nicotinamide mononucleotide (NMN) via an adenylylation reaction (Hashida et al. 2009).
NAD+-biosynthesis is crucial in multiple metabolic events and therefore a continuous synthesis of NAD+ is of vital importance to all cells. As mentioned previously in (Belenky et al. 2007), not only does NAD+ act as an coenzyme, whose concentration must be maintained to sustain basic cellular redox and energy metabolism, but also as substrate for NaM producing enzymes, particularly ADP-ribose transferase also known as poly(ADP-ribose)polymerase (PARP). The latter is the key enzyme in one of the major NAD+-consuming process, the poly(ADP-ribosyl)ation. This covalent post-translational protein modification, which utilizes NAD+ as substrate for NaM synthesis, has caught notable attention over decades in animal research (Briggs and Bent 2011; Bürkle 2001). In animals, poly(ADP-ribosyl)ation participates in several cellular processes such as DNA-repair, DNA-replication, regulation of cell cycle and in maintaining genomic stability (Sallmann et al. 2000; Trucco et al. 1998). During the reaction catalyzed by this enzyme branched ADP-ribose polymers are formed and attached to glutamate residues on specific protein receptors (Adams-Phillips et al. 2010; Hunt and Gray 2009; Hunt et al. 2004). The modification is reversed by poly(ADP-ribose) glycohydrolase (PARG), which hydrolyzes poly(ADP-ribose) polymers to form free ADP-riboses (Briggs and Bent 2011; Meyer et al. 2006). Due to the fact that poly(ADP-ribosyl)ation plays a vital role in numerous cellular processes and also affect heart attack, ischemia, Alzheimer and sensitivity of therapeutic cancer treatment reagents (Andrabi et al. 2006; Briggs and Bent 2011; Meyer et al. 2006), the development of PARP inhibitors has been a priority for pharmaceutical companies (Briggs and Bent 2011; Heeres and Hergenrother 2007).
By contrast, very few reports in plant systems have been published to date (see Briggs and Bent 2011; Jia et al. 2013; Pétriacq et al. 2012; Schulz et al. 2012). The first reports on the function of PARP in plants appeared in the late 70s (Payne and Bal 1976; Whitby and Whish 1977), where poly(ADP-ribosyl)ation was detected in nuclei of both germinated and non-germinated Allium cepa seeds as well as in isolated nuclei from root tips of Triticum aestivum. Only relatively recently, however, was the role of PARP during biotic and abiotic stress responses revealed (Amor et al. 1998; de Block et al. 2005; Vanderauwera et al. 2007). Furthermore, molecular genetic approaches were only attempted very recently (Jia et al. 2013; Rissel et al. 2014; Schulz et al. 2012). These studies suggested the participation of PARP in stress response, activation of non-homologous end-joining repair mechanism and seed development. It is important to note however that the first study via use of non-specific chemical and genetic inhibition did not dissect the roles of the specific isoforms. Furthermore, a detailed basic molecular characterization of the expression and localization of the independent isoforms is currently lacking. Here we, therefore, investigated expression and localization for the three Arabidopsis isoforms of PARP. In addition we physiologically characterized parp1, parp2 and parp3 mutants at the level of germination rate, root growth, photosynthetic performance, reproductive development and shoot. The results presented are discussed in terms of the consequences of the mutations on NAD+ metabolism, metabolic fluxes and whole plant vegetative and reproductive phenotypes suggesting potential isoform-specific non-nuclear roles for the PARPs in Arabidopsis thaliana.
Results
Expression patterns of PARP1, PARP2 and PARP3 in arabidopsis
Despite the fact that previous studies have focused on the biological function of PARP in Arabidopsis (Jia et al. 2013; Rissel et al. 2014; Schulz et al. 2012), surprisingly they did not carry out basic expression analysis of the three PARP genes encoded in the Arabidopsis genome (indeed this information is currently available only for PARP3). Scanning the Bio-Analytic Resource for Plant Biology website (BAR, bar.utoronto.ca, Toufighi et al. 2005), suggests high expression of PARP1 and PARP2 in the shoot apex, young siliques between stage three and five, and closed and open flowers. Moreover these two genes also appear to be expressed in dry seeds, young seedlings and late seedlings. By contrast, PARP3 is suggested to be expressed only in dry seeds and seeds of mature siliques (Jia et al. 2013; Rissel et al. 2014; Schulz et al. 2012, 2014). Our own data in leaves, roots and seeds were very much in support of this with PARP3 barely being expressed in leaves and roots but massively so in seeds with the other isoforms displaying contrasting expression patterns (Table 1).
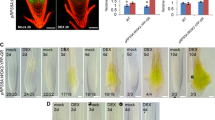
In order to investigate organ and cell specificity expression in more detail, the PARP1, PARP2 and PARP3 5′ upstream regions (2360 base pair, 2047 base pair and 1833 base pair, respectively) were fused to the β-glucuronidase (GUS) gene. In dry seeds GUS expression was only visible in plants bearing the GUS gene under the control of the PARP3 5′ upstream region (Fig. 1a), however in imbibed seeds and young seedlings staining was visible when GUS expression was regulated by the PARP1 and PARP3 promoters (Fig. 1b, c). In addition, in older seedlings (21 day old), GUS staining was highest in pPARP1::GUS and pPARP3::GUS lines (Fig. 1d). According to our GUS lines PARP1 was the only isoform highly expressed in the stamen of the open flower (Fig. 1e). None of the isoforms were expressed in siliques (Fig. 1f), whilst PARP1, PARP2 and PARP3 were all expressed in roots (Fig. 1g). These results thus largely mirror those described found on the Bio-Analytic Resource for Plant Biology website (BAR, bar.utoronto.ca, Toufighi et al. 2005), however, since GUS reporter lines prone to lacking regulatory elements in the 5′ upstream region fused to the reporter gene, we wanted to compare the expression pattern with RNA in situ hybridizations on various tissue sections (Fig. 2). While PARP1 was only present at very low concentrations at the shoot apex of vegetative meristems from long day and short day grown plants and in heart, torpedo and U-stage embryos (Fig. 2a, d, g, j, m, p), PARP2 transcripts were highly abundant in vegetative as well as inflorescence apices (Fig. 2b, e). Additionally, we found PARP2 to be expressed in heart and torpedo stage embryos (Fig. 2k, m). Hybridization with the PARP3 probe on the same tissue led to very weak to no staining in both the shoot and the root apical meristem as well as to a weaker staining in young vascular tissue of U-stage embryos (Fig. 2r). For each transcript hybridization with a control probe in the sense orientation gave no signal (Fig. 2s–u).
RNA in situ hybridization using specific probes for the PARP genes on longitudinal sections through vegetative apices (a–c long day; d–f short day), inflorescence apices (g–i), embryo at various stages (j–l heart stage; m–o torpedo stage; p–r U stage) and sense control in long day vegetative apices (s–u)
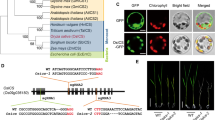
Based on the sequence information informatic approaches (BAR, bar.utoronto.ca, Toufighi et al. 2005), predict PARP1, to be localized to nucleus only, whereas PARP2 predicted to be present in the nucleus, mitochondria and chloroplasts and PARP3 could be highly expressed in the nucleus and slightly also in the cytoplasm. Given that such predictions have been demonstrated to be subject to error we cloned the full-length cDNAs of all three PARP genes and expressed them in-frame with a C-terminal GFP encoding gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter (p35S). These constructs were subsequently transformed into Arabidopsis and fluorescence patterns were compared, by confocal microscopy, to the chloroplast auto-fluorescence, a nuclear control and the mitochondrial marker mitotracker within the given cell. When expressed in cell suspension culture a nuclear location of all three PARP gene products like that recently demonstrated by Song et al. (2015). However, protoplasts transformed with p35S::PARP1::GFP exhibited fluorescence, which was not restricted only to the nucleus, but also visible in the chloroplasts (Supplementary Fig. 1). Inspection of plants transformed with p35S::PARP2::GFP displayed strong fluorescence signals in both chloroplasts (Fig. 3D1–D3) and mitochondria ((Supplementary Fig. 1). In accordance to recent results described by (Rissel et al. 2014); PARP3 was clearly localised at high abundance in the nuclei (Supplementary Fig. 1. Additionally, we observed a partial localization of PARP3 in cytosol (Supplementary Fig. 1. Thus, whereas animal PARPs have all been identified as nuclear proteins (Ame et al. 2004; Andrabi et al. 2006), the subcellular localization of PARPs homologues in Arabidopsis leaf protoplast is not exclusively nuclear and varies slightly between the isoforms (Supplementary Fig. 1).
Expression of PARP1, PARP2 and PARP3 in Arabidopsis protoplast. p35S::PARP::GFP constructs were transiently expressed in protoplast derived from 3-week-old Col-0 rosette leaves grown in long day (16 h photoperiod) to detect the subcellular localization of the PARPs. With confocal microscope GFP is visualized in green, red indicates chlorophyll auto-fluorescence in chloroplasts
Isolation (and complementation) of knockdown mutants of PARP1, PARP2 and PARP3
The finding that the various PARP isoforms have distinctive expression patterns indicates that they may have functionally divergent roles depending on the tissue and developmental stage, we next isolated knockdown mutants for PARP1, PARP2 and PARP3. For this purpose independent Arabidopsis lines that contained T-DNA elements inserted into the respective PARP genes were ordered from Nottingham Arabidopsis Stock Centre (NASC, Loughborough-UK). Homozygous T-DNA insertion lines for each PARP were confirmed by genomic PCR and assigned as parp1, parp2, and parp3 (Fig. 4a–d). RT-PCR was next carried out to check for expression of the full-length parp1, parp2, and parp3 transcripts (Fig. 4e–g). These studies revealed that the insertional mutants were knockouts for the allele targeted. Following this, the level of transcript of the other PARP genes as well as the expression of NADS, NaMNAT, SRT1 and SRT2 of the NAD+-biosynthesis pathway and the expression of mitochondrial malate dehydrogenase (MDH), fumarase (FUM) and NADP-dependent isocitrate dehydrogenase (NADP ICDH) in leaf tissue were investigated in each of the mutants (Fig. 5). The PARP1, PARP2 and PARP3 transcripts were non-determinant in parp1, parp2 and parp3 mutants, respectively (Fig. 5). The parp1 mutant was characterized by a strong decreased expression of NaMNAT and between two- and threefold increased expression of MDH, FUM and NADP ICDH. The parp2 mutant was additionally characterized by elevated expression of NADS and SRT1 and SRT2 but no notable difference in the expression of the examined TCA cycle transcripts. The parp3 mutant displayed a minor decrease in the expression of PARP2, but a compensatory increase in the expression of PARP1 by almost threefold (Fig. 4). Similar to the other two T-DNA lines, parp3 showed an increased expression of NADS, though only parp2 was characterized as significant. Like parp1 line, parp3 displayed between two and threefold increased expression of MDH, FUM and NADP ICDH (Fig. 5). Each of the knockout mutants was complemented by the expression of the targeted gene under the control of the 35S promoter.
Agarose gel electrophoresis separation of genomic PCR performed in (a) parp1, (b) parp2 and (c) parp3 T-DNA insertion lines. PCR product with gene specific forward/reverse primers (A), PCR product with right border gene specific primer in combination with T-DNA insertion specific primer (B), 1kb gene ladder (L) was used to determine the size of PCR product (d). Genomic PCR was performed on parp1, parp2, parp3 and wild type Col-0 with primer sets amplifying the genes PARP1, PARP2, PARP3. (e–g) Agarose gel electrophoresis of amplified PARP full length cDNAs in Col-0 and parp mutants. Leaves of 3-week-old Col-0 and parp lines grown in long day (16 h photoperiod) were harvested for RNA isolation and cDNA synthesis. Full length cDNA PCR was performed to determine the expression level of the following genes: (e) PARP1, (f) PARP2 and (g) PARP3 in wild type Col-0, parp1, parp2 and parp3
NAD+ hydrolase enzymatic activity via GC–MS analysis of nicotinamide (NaM) content in wild type Col-0 and the parp lines. Leaf whole cells as well as nuclei of plants grown in long day were isolated and extracts were desalted. Enzymatic assay was performed with addition of 1 mM NAD+ as substrate (without substrate in blank assays) and 60 min incubation time. Relative NaM content was normalized by the amount of NaM in blank assay, ribitol, fresh weight and incubation time. Values are mean ± SE of five to six biological replicates. Asterisks represent values determined by Student’s t test to be significantly different (p < 0.05) from Col-0. N.D. indicates not determined
Development of a method to assay the total cellular and nuclear NAD hydrolase activities
As mentioned above, PARP uses NAD+ as substrate in the salvage pathway to produce NaM via poly(ADP-ribosyl)ation. In order to investigate whether the decreases in PARP transcript resulted in similar changes at the enzyme activity level, an assay was established to determine the production of NaM in whole cell extracts as well as nuclei preparations from leaf tissue. This approach was taken given the difficulty in detecting poly(ADP-ribose) and as a means to discriminate PARP activity from that of the parallel reaction catalyzed by sirtuin (König et al. 2014) and the similar reaction catalyzed by nudix enyzmes (Hashida et al. 2009), although it is important to note that it may not discriminate PARP from other potential NAD+ hydrolase activities. Thus we are only able to quantify the total NAD+ hydrolase activity by this approach. For this purpose fresh Arabidopsis rosette leaf tissue was ground and incubated in a simple isolation buffer as described previously by (Folta and Kaufman 2006) prior to filtering through two layers of miracloth. To isolate nuclei, the filtered suspension was then lysed and centrifuged to enrich for nuclei. To remove metabolites or peptides, which could interfere with the reaction, a desalting step was carried out. The enzymatic assay was then performed with a 1 mM NAD+-containing enzymatic reaction buffer for 60 min at room temperature and stopped by addition of ice-cold trichloroacetic acid. The NaM produced in the end-point assay was subsequently extracted and measured by gas chromatography-mass spectrometry (GC–MS) as detailed in the “Materials and methods” section. Given that this was an assay novel to us, before analyzing the mutant lines we verified that it was linear with respect to both time and protein concentration (data not shown). The cellular NAD+ hydrolysis activity of wild type Arabidopsis observed (Fig. 6) was similar to those previously reported for PARP in mammals and maize (Grube and Bürkle 1992; Tian et al. 1999). A reduced activity was observed for the parp1 and parp2 mutants in both extracts of whole leaf extracts and nuclei preparations, although the reduction was statistically significant only for the nuclei (Fig. 7). Expectedly, the activity in parp3 mutant was at wild type level, since PARP3 is not highly expressed in vegetative tissue, however importantly the enhanced expression of PARP1 in this line did not result in an increased overall PARP activity.
PARP enzymatic assay calibration curve. Leaf nuclei of Col-0 grown in long day (16 h photoperiod) were isolated. Filled black circles represent enzymatic assay performed with addition of 1 mM NAD+ as substrate with different incubation time (20-40-80-120-180 min). Filled red circles represent data of enzymatic assay performed with addition of different substrate concentration (0.5-1-3 mM NAD+) and 120 min incubation time
NAD+ hydrolase enzymatic activity via GC–MS analysis of nicotinamide (NaM) content in wild type Col-0 and the parp lines. Leaf whole cells as well as nuclei of plants grown in long day were isolated and extracts were desalted. Enzymatic assay was performed with addition of 1 mM NAD+ as substrate (without substrate in blank assays) and 60 min incubation time. Relative NaM content was normalized by the amount of NaM in blank assay, ribitol, fresh weight and incubation time. Values are mean ± SE of five to six biological replicates. Asterisks represent values determined by Student’s t test to be significantly different (p < 0.05) from Col-0
Characteristics of seed germination and root growth
To study the impact of PARP inactivation on plant growth we next compared seed germination of the mutants with that of the wild type in the presence of sucrose. Using freshly harvested seeds we observed that on half-strength Murashige and Skoog (MS) agar with 1 % sucrose parp3 started to germinate after 3 days, thus earlier than Col-0, parp1 and parp2 mutants (Fig. 8a). While after 4 days parp2 and parp3 lines continued to show a higher germination rate compared to wild type, the final germination rate of parp3 was similar to Col-0, whereas the other two insertion lines revealed an overall slower and slightly reduced germination rate (Fig. 8a). A similar germination rate was observed on half-strength MS agar without 1 % sucrose supplement, indicating, that PARP3 deficient plants were capable of sugar independent germination. More importantly by using complemented mutant lines we could observe the rescue of the wild type phenotype (Fig. 8a).
Germination rate and root elongation rate of parp mutant, complemented lines cPARP and wild type Col-0. Sterilized seeds were grown on half-strength Murashige and Skoog (MS) agar supplemented with 1 % sucrose under long day condition (16 h photoperiod). a Germination rate is expressed as total germination in percentage ±SE of three repetitions and b root length is expressed as mean ± SE. Asterisks represent values determined by Student’s t test to be significantly different (p < 0.05 and p < 0.01) from Col-0. n.g. no germination
As a next experiment the root development of homozygous lines was determined on 2MS medium (Fig. 8b). A higher root elongation rate was generally observed in all mutant lines in comparison to wild type. Although parp1 was only significantly higher than Col-0 after 4 days, parp2 and parp3 were significantly higher than Col-0 throughout the experiment (Fig. 8b). Complemented mutant lines cPARPs, however, displayed a reduced root elongation compared to their respective mutants. Notably this result is somewhat different between the lines indicating functionally non-redundant roles for the isoforms.
Metabolic alterations in leaves of parp mutants
To further characterize the roles of the independent PARP isoforms we next performed metabolite profiling in leaves of the mutants. Despite lacking changes in total chlorophyll content, the chlorophyll a/b ratio was reduced in parp2 and increased in parp1, whereas no differences between wild type and parp3 were detected. We further investigated whether these changes in chlorophyll might be associated with photosynthetic activity observing minor yet significant reductions in the PSII maximum efficiency after dark adaptation (F v/F m) as well as in electron transport rate (ETR; not significant in parp1) compared to Col-0 (Fig. 9). It is worth mentioning that the changes in F v/F m and ETR were minor in plants of all genotypes, typically indicative of a lack of major stress and rather associated with general changes in metabolism (Essemine et al. 2012). Additionally, gas exchange was measured directly in 4-week-old plants growing under long day condition, under photon flux densities (PFDs) that ranged from 25 to 1000 μmol m−2 s−1. All parp mutants exhibited unaltered assimilation rates, stomatal conductance, intracellular and ambient CO2 ration (C i/C a) and transpiration rates (Fig. 10). Furthermore, we measured the rate of dark respiration using via infrared gas-exchange analyses (Fig. 11). These measurements revealed that in all parp mutants there is a tendency of reduction in dark respiration, and although this was not significant in any of the genotypes, the alteration in parp2 was higher than in parp1 and parp3 (Fig. 11). When taking together, alongside the results of in F v/F m and ETR, these data strongly suggest that photosynthetic machinery is not compromised in the absence of any individual parp isozymes.
Measurement of chlorophyll fluorescence and the efficiency of electron transport in dark adapted leaves of parp lines in comparison to wild type Col-0. Leaves of 4-week-old plants grown under long day (16 h photoperiod) were clamped in darkness for 10 min prior applying a light source to determine the photochemical efficiency of photosystem II [ratio of maximal variable fluorescence to maximum yield of fluorescence (Fv/Fm)] and the electron transport rate (ETR). Values are mean ± SE of seven biological replicates and asterisks represent values determined by Student’s t test to be significantly different (p < 0.05) from Col-0
Effect of decreased PARP activity on photosynthetic parameters. 4-Weeks-old in long day (16 h photoperiod) grown parp lines and wild type were dark adapted 30 min prior exposure to light sources of various intensities. Stomatal conductance, assimilation rate and stomatal transpiration rate were determined. Values are mean ± SE of 4–5 biological replicates and asterisks represent values determined by student’s t test to be significantly different (p < 0.05) from Col-0
Effect of decreased PARP activity on respiration of 4-week-old plants. parp lines and wild type plants grown in long day (16 h photoperiod) were dark adapted 30 min prior exposure to light source. Values are mean ± SE of 4–5 biological replicates and asterisks represent values determined by Student’s t test to be significantly different (p < 0.05) from Col-0
To further elucidate the role of PARPs in Arabidopsis thaliana we next used an established GC–MS platform that affords good coverage of the major metabolites of primary metabolism (Fernie et al. 2004; Lisec et al. 2006) to quantify the relative metabolite levels in leaf samples of parp mutants in order to confirm the suggested alteration in plant metabolism. These studies revealed considerable changes in the levels of a wide range of organic acids, amino acids and sugars (Table 2). In order to characterize changes in the metabolome Tukeys tests were employed wherein the wild type was independently compared to each mutant and it´s respective complemented line we describe a change as significant only when it is altered in the mutant and not in the complemented line or when it is altered in the mutant and the complemented line is significantly different from the mutant and reverting its change. The levels of many amino acids were reduced significantly in one or more of the mutant lines for example alanine and GABA in parp1, isoleucine and ornithine in parp2 and parp3 and glycine, phenylalanine and tyrosine in parp2 alone. By contrast, aspartate levels were increased in parp1 and parp3 and glutamine in parp2 and parp3.
The levels of malate and dehydroascorbate were increased in parp-1 whilst succinate was increased in parp-1. By contrast, glycerate was decreased in parp-1 and parp-3, lactate was decreased in parp-1 and parp-2 and pyroglutamate was increased in all lines. Threonate increased in parp-1 but decreased in parp-2. Furthermore, an increase in Glu and Suc content in parp-1 and parp-3, although not in was observed. Conversely the levels of fructose, raffinose and trehalose were decreased in parp-2, whilst levels of the latter increased in parp-3 and minor alterations in sugar alcohols and polyamines were also apparent.
We next directly evaluated the rate of light respiration in the mutant lines. For this purpose we recorded the evolution of 14CO2 following the incubation of leaf discs in positional-labeled 14C-glucose molecules in order to assess the relative rate of flux through the oxidative pentose phosphate pathway (OPPP) and the TCA cycle (Fig. 12). For this leaf discs were incubated in the light and supplied with [1-14C]-Glc, [2-14C]-Glc, [3,4-14C]-Glc, or [6-14C]-Glc over a period of 6 h. During this time the 14CO2 evolved was collected at hourly intervals. Carbon dioxide can be released from the C1 position, and to a lesser extent the C2 position, by the action of enzymes that are not associated with mitochondrial respiration. On the other hand carbon dioxide released from the C3,4 positions of glucose are directly associated with the activity of enzymes connected to the mitochondrial respiration (Nunes-Nesi et al. 2007). Thus, the ratio of carbon dioxide released from C3,4 to C1 positions of glucose provides a strong indication of the relative rate of the TCA cycle in regard to other processes of carbohydrate oxidation. An interesting pattern was observed when the relative 14CO2 release of the mutant and wild type lines is compared for the various fed substrates regardless of the labeled position in the substrate. For instance following either [1-14C]-(Fig. 12a) or [2-14C]-labeling (Fig. 12b), CO2 release of parp1 was higher than wild type, while for parp2 plants a higher CO2 release for [1-14C] and a similar CO2 release for [2-14C] was observed. In addition parp3 plants showed a reduced CO2 release for [1-14C] and an increased CO2 release for [2-14C]. The CO2 release from the position [3,4-14C] also revealed an interesting pattern (Fig. 12c) with both parp1 and parp3 displayed increased evolution compared to wild type, while parp2 was invariant from wild type. Additionally, the CO2 release from the C6 position (Fig. 12d) of parp1 and parp2 are higher than wild type, while that from parp3 was similar to wild type. There was, furthermore, a shift in the ratio of CO2 evolution from the various labelled substrates, with the relative release from the C3,4 positions in relation to C1 in comparison to wild type plants. Ratios were as follows: wild type = 0.54 ± 0.07; parp1 = 0.37 ± 0.04; parp2 = 0.61 ± 0.06, and parp3 = 0.94 ± 0.15). These data suggested a tendency of a higher proportion of carbohydrate oxidation performed by the TCA cycle in parp2 and parp3 plants (significant in parp3).
Evolution of 14CO2 in wild type Col-0 and parp mutants grown in long day (16 h photoperiod). Leaf discs of 4-week-old plants were isolated and incubated in MES-KOH solution (pH 6.5) supplemented with 2.32 kBq mL−1 of a D-[1-14C]-, b D-[2-14C], c D-[3,4-14C]- or d D-[6-14C]-glucose. The 14CO2 release was captured hourly and quantified by liquid scintillation counting. Values are mean ± SE of three biological replicates each d-glucose labelling. Asterisk represent values determined by Student’s t test to be significantly different (p < 0.05) from Col-0
Since the poly(ADP-ribosyl)ation activity might also be associated with the consumption of oxidized coenzyme NAD+, it is reasonable to anticipate that reduction in the activity of PARP enzymes might affects the redox balance in the knockout plants. Therefore, we decided to determine the levels of pyridine dinucleotides in the leaves of wild type and mutant plants (Fig. 13). An increase in the level of NAD+ was observed in parp1) while no significant differences were observed for parp2 and parp-3 plants (Fig. 13a). Although a tendency of increased NADH content was observed for all lines, no significant changes were detected in the content of this dinucleotide (Fig. 13b). In addition only in parp3 was a decreased NADH/NAD+ ratio observed (Fig. 13c). Significant increases in the content of NADP+ and NADPH were, however, observed for parp1 and parp3 plants (Fig. 13d, e). Despite these changes there were no significant alterations in the NADPH/NADP+ ratio (Fig. 13f).
Pyridine nucleotide levels and ratios in leaves of WT Col-0 and parp lines. Leaf materials of 3-week-old plants grown in long day (16 h photoperiod) were harvested in the middle of the light period and extracted to determine the redox status of the parp lines in comparison to wild type. Detected content of NAD(H) and NADP(H) are given in nmol g−1 FW with NAD+ content (a), NADH content (b), NADH/NAD+ ratio (c), NADP+ content (d), NADPH content (e) and NADPH/NADP+ ratio (f). Values are mean ± SE of three to eight biological replicates. Asterisk represent values determined by the Student’s t test to be significantly different (p < 0.05) from wild type
Metabolic profile in roots of parp mutants
To follow the repertoire of metabolic changes that might explain the reasons behind the root phenotype in parp mutants we have also performed an extensive metabolite profiling in roots of 4-week-old Arabidopsis plants grown under long day conditions (Table 3). Changes were statistically assessed in the same manner as above for the shoots there were no changes in protein, nitrate or total amino acid levels nor in sucrose or fructose but the levels of glucose were reduced in parp-1 and parp-2 whilst the levels of starch were decreased in parp-1 and parp-3. Perhaps surprisingly, there were very few changes in the levels of metabolites determined by GC–MS in either parp-1 or parp-2. The parp-1 mutant displayed increases in asparagine and beta-alanine, whilst parp-2 was unaltered whilst parp-3 displayed the decrease in glucose we observed by spectrophotometric methods. By contrast, parp-3 displayed decreased levels of intermediates of the photorespiratory pathway namely glycine and serine as well as glycerate and glycolate as well as the stress amino acids GABA and proline and the TCA cycle intermediates malate and succinate. Conversely, the levels of methionine and phenylalanine as well as those of phosphorate were increased in parp-3. In addition, the levels of raffinose, xylose, galactionol, myo-inositol, threitol and urea were all decreased in parp-3.
Discussion
Here we demonstrate that the isoforms of PARP play non-redundant roles in Arabidopsis under non-stressed conditions. Although the metabolic role of PARP has been the subject of considerable research in mammals where for example roles have been documented for PARP1 in mitochondrial metabolism and PARP2 in pancreatic function and whole body energy expenditure (Bai et al. 2011a, b; Luo et al. 2001), that in plants is much less well characterized. Indeed, until recently such studies were limited to observations of correlative increases in PARP activity, pyridine nucleotide cycling and the nuclear localization of glutathione during exponential cell growth of Arabidopsis (Pellny et al. 2009) and the observation that lines silenced in PARP activity using RNAi exhibited a better energy use efficiency by reducing NAD+ breakdown (de Block et al. 2005). More recent studies of PARP function revealed a 30 % increase in NAD+ levels and considerable changes in the expression of genes associated with anthocyanin biosynthesis and a strong reduction of anthocyanin accumulation in stress conditions (Schulz et al. 2012). Furthermore a detailed study of the growth in PARP inhibited Arabidopsis plants revealed enhanced ATP and NAD+ levels but no clear increases in the levels of other primary metabolites leading the authors to conclude that any additional primary metabolites made following treatment were immediately utilized in the support of growth (Schulz et al. 2014). It is, however, important to note that these studies were either indirect or reliant on the use of chemical inhibitor studies or an RNAi approach targeting all PARP isoforms and as such cannot discriminate between the individual roles of the different isoforms of the enzyme. By contrast, a recent study has comprehensively characterized the function of the sirtuin SRT2 which functions as a mitochondrial lysine deacetylase whilst simultaneously producing one molecule of nicotinamide (König et al. 2014). The study suggested the role of SRT2 in the regulation of specific proteins, such as ATP synthase and ATP/ADP carriers and thus its involvement in mitochondrial energy metabolism.
Here we show that the PARPs have a similar yet distinct metabolic function to SRT2. In addition we performed a detailed analysis of the expression profiles of the PARPs as well as studying the localization of their gene products and the distribution of PARP activity. When taken together these combined data reveal novel roles for PARP genes under non-stress conditions in addition to the better characterized roles in DNA repair and defense response (Amor et al. 1998; Babiychuk et al. 2001; Berglund and Ohlsson 1995; Rissel et al. 2014; Song et al. 2015) Interestingly, PARP1 and PARP3 appear to have more important metabolic roles, which is perhaps consistent with the recent observation that PARP2 is the dominant isoform for DNA damage repair and immune responses (Song et al. 2015) That PARP3 displays such dramatic metabolic changes in leaves and roots is rather surprising in light of its expression pattern. We cannot at present explain the precise mechanism underlying this, however, two possibilities warrant commenting upon. The first and one we favor is that this effect is somehow carried over from effects in the seed wherein PARP3 is massively expressed whilst the second is that modulations of this very low expression is sufficient for the phenotype (providing that the location is correct—see below). It is additionally important to note that changes in the root metabolism of the PARP3 mutants could be the consequence of its restricted root growth and thus be indirect effects. That said in the absence of inducible mutants it is very difficult to tease apart the causality of the metabo- to phenotype relationship.
Expression and localization of the various PARP isoforms and molecular characterization of T-DNA insertional knockdown mutants
The vast majority of work on the roles of PARP in plants have focused on its role in DNA repair or, in the case of the few metabolic studies, have been either indirect (Pellny et al. 2009), or carried out using chemical inhibitors which are likely to inhibit all isoforms to a similar extent (Schulz et al. 2012, 2014). For this reason as a first step to characterize the individual non-redundant roles of the PARP isoforms we evaluated their levels of expression both by the use of promoter GUS fusions (Fig. 1) and by RNA in situ hybridization experiments (Fig. 2). These results suggest that PARP2 has a distinctive expression pattern from the other isoforms being not or only weakly expressed in seeds but exhibiting high expression in vegetative meristems, whilst PARP1 was the only isoform expressed in floral tissues and as such suggest at least partially non-redundant functions. However, that said, there is considerable overlap between PARP1 and PARP3 expression and all isoforms are expressed in the root where they may play redundant functions. Intriguingly, however, by contrast to a previous report (Boltz et al. 2014), our expression analysis did not support a negative correlation between the expression of PARP1 and PARP2, suggesting that this may well not be a direct effect.
We additionally assessed the subcellular localization of the isoforms using p35S::PARP::GFP fusion constructs (Fig. 3). These data were largely in agreement with sequence based predictions (BAR, bar.utoronto.ca, Toufighi et al. 2005) in suggesting nuclear localizations (although we could not see this for PARP2) but also additional extra-nuclear localizations with signals for PARP1 also present in the chloroplast, PARP2 the chloroplast and mitochondria and PARP3 in the cytosol. Finally, we analyzed the expression of PARP genes in the knockdown lines: parp1, parp2 and parp3 (Fig. 4). A complete lack of expression was observed as expected when analyzing the PARP gene in its respective T-DNA line, furthermore a complementary increase of PARP1 was seen in the parp-3 mutant. We next evaluated the total cellular NAD+ hydrolase activity in whole leaf and nuclear extracts of the mutants using an end-point assay determination of nicotinamide production over time. These assays revealed mild yet non-significant decreases of NAD+ hydrolase activity in whole leaves of the parp1 and parp2 mutants and significant decreases (of up to 50 %), in the nuclear activities in these mutants (Fig. 7), with no change in the level of the parp3 mutant consistent with the negligible expression of this isoform in leaf tissue. Interestingly, despite the clear indication of extra-nuclear localization of PARP proteins both from our GFP studies and, at least in the leaf, the NAD+ hydrolase activity is clearly predominantly localized in the nucleus. This probably goes some way to explaining why PARPs have not been reported to date in organellar proteome surveys (suba.plantenergy.uwa.edu.au), however, it is important to note that there is plenty of anecdotal evidence that low abundance proteins are below the level of detection of such methods.
Effect of knockout of PARP isoforms on growth phenotypes
The first obvious phenotype of the parp mutants was that all three mutants germinated faster even though the final germination efficiency was invariant from wild type Previous studies have already proposed the relationship of PARPs in protecting the seeds from genotoxic stress (Hunt and Gray 2009; Hunt et al. 2007) and although no direct evidence has yet been provided for the involvement of PARP in seed dormancy it was recently shown that PARP3 deficient plants do not maintain germination rates after storage (Rissel et al. 2014). The role of NAD+ metabolism in regulating seed dormancy has been much discussed with seed poly(ADP-ribose) levels correlating well with sensitivity of germination to the DNA damaging agent methyl methanesulphonate, whilst the level of NAD+ appears to affect the depth of dormancy potentially by enhancing abscisic acid (ABA) synthesis (Hunt and Gray 2009). Consistent with this hypothesis is the finding that genes knocked out in the nicotinamidase gene NIC2 display retarded germination and were hypersensitive to addition of nicotinamide or ABA suggesting that parp activity had been impaired (Hunt et al. 2007). Our results suggest that although parp3 appears to be considerably more highly expressed in the seed than the other isoforms, it has very little influence on the rate of germination. The rate of root elongation was, by contrast, more dramatically altered with all three mutants but with parp-2 and parp-3 especially displaying considerably accelerated root growth. Results of GC–MS based metabolite profiling, however, revealed that no metabolic changes were observed in all three mutants. Arguably of highest interest is the fact that the photorespiratory metabolites glycerate, glycine, glycolate and serine all decreased in parp3). This is of high interest given the clearly established link between NAD+ metabolism, photorespiration and nitrogen assimilation in folial tissues (Bauwe et al. 2010) and our recent finding of important metabolic roles for these enzymes in Arabidopsis roots (Nunes-Nesi et al. 2014).
Effect of downregulation of PARP isoforms on leaf metabolism and function
The canonical marker for chemical PARP inhibition is the accumulation of NAD+ (de Block et al. 2005). Interestingly, the level of this metabolite was only significantly increased in leaves of the parp1 mutant, with the NADH/NAD+ ratio even being higher in the parp3 mutant. Conversely the NADP+ and NADPH levels both increased in parp1 and parp3 mutants with the NADPH/NADP+ ration being unaltered in both mutants. These results thus suggest that the increase in NAD+ observed following chemical inhibition of PARP most likely largely reflects a modulation of the PARP1 isoform. A considerable amount of further metabolic changes were observed in the parp mutants most notably in parp1 (which displayed significant alterations in the contents of 15 metabolites) but also in parp2 and parp3 (which displayed 14 and 13 changes, respectively). Despite the large degree of change in metabolite levels intriguingly only the increase pyroglutamate was observed in all lines. There are however multiple examples of changes that were conserved between two out of the three lines with parp1 and parp3 sharing an additional five changes, whereas parp1- and -2 shared an additional three and parp2 and -3 shared three additional changes (Fig. 13), with the remaining changes being unique to one of the isoforms. When comparing these changes to those previously reported for other mutants associated with NAD+ metabolism (König et al. 2014; Schippers et al. 2008), some interesting commonalities are apparent. For example gluatamine, which was significantly increased in two of the three parp mutants was also increased in plants mutated in the parallel sirtuin activity but changes in sugars, organic acids and the other amino acids were dissimilar between the srt2 mutant and the parp mutants (compare data from this study with that from König et al. 2014). The changes in at least parp1 and parp3 were however considerably more similar to those in the old5 genotype which is a mutant of quinolate synthase, a key enzyme in the de novo synthesis of NAD+. For example the changes in sucrose, malate and aspartate were highly similar in parp1, parp3 and old5 (compare data from this study with that from Schippers et al. 2008). Whilst there is considerable overlap between the changes in the various mutants associated with NAD+ biosynthesis there was surprisingly little commonality in the parp mutants metabolic phenotype and that induced following chemical inhibition by 3-Methoxybenzamide (3 MB) (Schulz et al. 2014). Moreover, the changes in the mutants are quite distinctive from those observed in the roots tending to be quite similar between the lines despite the fact that PARP3 expression is considerably lower than the other isoforms in this tissue. Indeed the changes are largely consistent with those of the gas exchange measurements which revealed no difference in the rate of photosynthesis but a mild yet insignificant reduction in the rate of respiration. The majority of the changes are similar to those observed previously following the manipulation of mitochondrial metabolism. For example, the up regulation of dehydroascorbate mirrors that observed following the up-regulation of the galacto-1, 4-lactone dehydrogenase donation to the mitochondrial electron transport chain (Nunes-Nesi et al. 2005). Finally, parp-3 roots are characterized by massive and conserved changes in the levels of all detected photorespiratory intermediates this finding is highly interesting particularly given current models of the interaction of NAD+ with photorespiration (Bauwe et al. 2010), and our recent findings that the majority of this pathway is fully operational in heterotrophic tissues (Nunes-Nesi et al. 2014).
In summary, this study provides the first detailed molecular characterization of the PARP family in Arabidopsis and comprehensively characterizes the non-nuclear metabolic and physiological role of the independent isoforms under non-stress conditions. These studies provide support for current models of the linkage between NAD+ metabolism, the TCA cycle and associated mitochondrial metabolism and photorespiration revealing an role for them in the co-ordination of central metabolism. Given that the metabolic consequences of PARP deficiency were quite distinctive from that following the inhibition of the parallel reaction catalyzed by sirtuin (König et al. 2014), it is apparent that these proteins have differential roles in plant function with the PARPs seemingly more important with regard to metabolism. It is additionally intriguing that there is a significant increase in the levels of NADP(H) in the leaves of Parp1 and -3 mutants whilst PARP would have been anticipated to rather effect NAD+ levels. In this context it is additionally interesting to compare and contrast our data with that found on the overexpression of NAD+ kinase in rice and Arabidopsis (Takahara et al. 2010; Takahashi et al. 2009). The NAD+ kinase also had only marginal effects on NAD+ levels but considerably elevated NADP(H) levels and were characterized by elevated amino acid content but little change in the levels of most other metabolites measured here. These results are partially overlapping with those of the PARP1 and PARP3 mutants i.e. for those amino acids which increase although it is important to note that many of the changes are opposite. Thus studies looking at the role of NADP(H) in the regulation of the commonly changing amino acids would likely be a promising avenue for future research. Finally, the root and seed phenotypes of the parp mutants provide direct proof for the previous postulates of a role for PARP in these developmental processes with our data suggesting a possible link between enzymes of the photorespiratory pathway and root growth. We are thus able to conclude that PARP isoforms play partially overlapping but non-functionally redundant roles in Arabidopsis metabolism and have importance above and beyond that already inferred by testing their role with regard to oxidative stress. It seems likely that organelle specific overexpression of PARP activities would be a highly informative approach by which to gather further understanding of their compartment specific functions and should be a priority for future studies.
Materials and methods
Chemicals
All standard chemicals and enzymes were obtained from Fluka (Seelze, Germany), Merck (Darmstadt, Germany), Roche (Mannheim, Germany) and Sigma Aldrich (Deisenhofen, Germany), unless stated otherwise.
Plant material
All Arabidopsis thaliana plants used in this study were of the Columbia ecotype (Col-0). The poly(ADP-ribose)polymerase (PARP) T-DNA mutant lines parp1 (SAIL_632_D07), parp2 (SALK_111410) and parp3 (SAIL_1250_B03) were obtained from Nottingham Arabidopsis Stock Centre (University of Nottingham, UK). Homozygous mutant lines were isolated by PCR using primers specific for PARP1 (P1-N878-LP, 5′-ACTTAATTGTGGTTTGACGCC-3′ and P1-N878-BP, 5′-CCGTCTTCTAGTGTTTGAGCC-3′) in combination with the T-DNA left border primer (SAIL-LB 3.1, 5′-CATCTGAATTTCATAACCAATCTCG-3′) for parp1, or primers specific for PARP2 (P2-N611-LP, 5′-AATTAAAGATGGACATTCGCG-3′ and P2-N611-RP, 5′-CAGAGAGAACAGGATTGAACCC-3′) in combination with the T-DNA left border primer (SALK-LBb 1.3, 5′-ATTTTGCCGATTTCGGAAC-3′) for parp2, and primers specific for PARP3 (P3-N875-LP, 5′-GTGGTACGTCAAAGATGGTGG-3′ and P1-N878-BP, 5′-TTGCAACAAACGAATCAAGC-3′) in combination with the T-DNA left border primer (SAIL-LB 3.1) for parp3.
Seeds were surface-sterilized and imbibed for 2 days at 4 °C in the dark prior to plating on half-strength Murashige and Skoog (MS) media with 1 % sucrose. Seeds were subsequently germinated and grown at 20 °C under long-day conditions (16 h light/8 h dark) with 150 μmol m−2 s−1. For germination rate and root growth analysis seeds were sown on MS media with 1 % sucrose.
GUS reporter activity assay
DNA fragments of the AtPARP promoters were amplified from the genomic DNA of Col-0 using the primers 5′-CACCTAGACGTGTGTAAAGTAGGCAAAGTG-3′ and 5′-TTTCGTCTTCTTCTTCAGGAGAATAG-3′, 5′-CACCTTTATTCACCATTTCTCTGCTTCTC-3′ and 5′-TTCTCCGGTAAGAGACAATTACACA-3′, or 5′-CACCATCGGTAAATGTAGCCAATAAGAC-3′ and 5′-TGAGCAAACTCTTTGAACTGTATG-3′ for PARP1, PARP2 or PARP3, respectively. The PCR product was recombined into pENTR/SD/D/TOPO (Invitrogen) and subsequently transferred into pKGWFS7 vector using Gateway technology. Transformants were selected with 50 mg L−1 kanamycin and GUS activity was detected by stereo electron microscope (Leica MZ12.5 stereomicroscope with Leica DFC420 digital camera) after vacuum infiltration with 2 mM 5-bromo-4-chloro-3-indolyl-β-d-Glucuronide in 50 mM sodium phosphate (pH 7.0), containing 0.2 % TritonX-100. Samples were incubated overnight at 37 °C and cleared in 70 % ethanol.
Assaying GFP expression in protoplasts
Full length coding sequences of PARP genes was isolated from Col-0 using the primers 5′-CACCATGGCGAACAAGCTCAAAGTC-3′ and 5′-GTGCTTGTAGTTGAATTTGACTTGG-3′, 5′-CACCATGGCAAGCCCACATAAGC-3′ and 5′-TCTCTTGTGCTTAAACCTTACTTTC-3′, or 5′-CACCATGAAGGTTCACGAGACAAGATC-3′ and 5′-CTCTGGTTCGACATCGACTATC-3′ for PARP1, PARP2 or PARP3 respectively. The PCR product was recombined into pENTR/SD/D/TOPO (Invitrogen) and subsequently transferred into pK7FWG2 vector using Gateway technology. Transformants were selected with 50 mg L−1 kanamycin. Plasmid construct p35S::NLS::GFP was used as control for nuclear localization. Protoplast isolation was performed by the TAPE-Arabidopsis-Sandwich protocol (described in Wu et al. 2009). Subcellular localization of PARP in plant compartments was detected under laser scanning confocal microscope (Leica TCS SP5) through fluorescence of GFP (green), mitotracker (blue) as well as chlorophyll auto—fluorescence (red).
RNA in situ hybridization
Plants were grown under long day (16 h light/8 h dark, 150 μmol m−2 s−1) at 22 °C and samples were collected from vegetative, reproductive plants and various embryonic stages. Fixation, dehydration and embedding into wax as well as probes synthesis and RNA in situ hybridization were performed as described in (Wahl et al. 2013). Histological sections were imaged with an Olympus BX-61 microscope equipped with a DC View III digital camera.
NAD hydrolase enzymatic assay
The assay was carried out with two different sets of extracts (nuclei only extract and whole cell extracts). The preparation of isolation buffer (consisting of 2 M hexylene glycol; 20 mM PIPES-KOH, pH 7.0; 10 mM MgCl2 and 5 mM β-mercaptoethanol) was prepared (exactly as described Folta and Kaufman 2006). Biological replicates of 0.5 g fresh Arabidopsis rosette leaves were harvested and grind in 10 mL of isolation buffer before filtering through two layers of miracloth. The filtered suspension was supplemented with 2 mL lysis buffer containing 10 % Triton-X100 in isolation buffer (for nuclei extract) and shook gently on ice or kept on ice in regards of whole cell extracts. After seven min the falcons were centrifuged at 4 °C for 10 min at 2000 g. Lastly, the pellet was dissolved in 1 mL storage buffer (consisting of 50 mM Tris–HCl, pH 7.8; 20 % glycerol; 5 mM MgCl2; 0.44 M sucrose and 10 mM β-mercaptoethanol). PARP activity assay was performed as described in (Tian et al. 1999). For each cell suspension and genotype, blank assay without addition of substrate was included. In parallel to the samples, nicotinamide standards (100–500 ng to 1–2 μg) were prepared and reduced to dryness in a Speedvac® overnight after addition of 60 μL ribitol. Extracted and dried samples above were extracted another time as well as derivatized with the protocol for metabolic profiling. Samples were put to run on the gas chromatography-mass spectrometry (GC–MS) with the nicotinamide (NaM)-standards, an Arabidopsis control and one blank control. The enzyme activities of PARP genes were determined by the amount of NaM via GC–MS method normalized to blank assay, internal standard, g fresh weight and incubation time.
Measurements of photosynthetic parameters
Leaf gas-exchange measurements were performed with an open-flow gas exchange system Li-Cor 6400 (Li-Cor Inc., Lincoln, NE, USA). Three-week-old plants grown in short day conditions and before bolting were used for determination of the photosynthetic parameters. Light response curves were carried out by varying the photosynthetic photon flux density (PPFD) from 1000 μmol m−2 s−1 to zero. The reference CO2 concentration was set at 400 μmol CO2 mol−1 air. All measurements were performed at 25 °C and the amount of blue light was set to 10 % PFD to optimize stomatal aperture. Additionally, using a dark-adaptation leaf clip, minimum (F 0) and maximum (F m) dark-adapted (30 min) fluorescence were measured, from which F v/F m ratio, in which F v = F m − F 0, was calculated. This ratio has been used as a measure of the potential photochemical efficiency of PSII.
Measurement of respiratory parameters
Dark respiration was measured using the same gas exchange system as defined above by adapting plants for 30 min in the dark to avoid light-enhanced dark respiration. Estimations of the TCA cycle flux on the basis of 14CO2 evolution were carried out following incubation of isolated leaf discs in 10 mM MES-KOH, pH 6.5, containing 2.32 KBq mL−1 of [1-14C]-, [2-14C]-,[3,4-14C]-, or [6-14C]-glucose. 14CO2 evolved was trapped in KOH and quantified by liquid scintillation counting. The results were interpreted following Ap Rees and Beevers (1960).
The 14C-labeling pattern of sucrose, starch, and other cellular compounds was determined by incubation of isolated leaf discs in 10 mM MES-KOH, pH 6.5, containing 10 μCi of [U-14C]-glucose at a PFD of 700 μmol m−2 s−1 in 22 °C for 2 h and subsequent fractionation was performed exactly as defined by Lytovchenko et al. (2002).
Metabolic profiling
Four-week-old plants were harvested for metabolite analysis. Chlorophyll, sucrose, starch, total protein, total amino acid and nitrate contents were determined as described in (Sienkiewicz-Porzucek et al. 2010), while NAD(H) and NADP(H) were determined as described previously (Schippers et al. 2008). Metabolite extraction for GC–MS was performed on the same samples as used for metabolite determination by spectrophotometric methods. The extraction, derivatization, standard addition, and sample injection and machine used were exactly as described by Lisec et al. (2006). Target metabolites were annotated using Chroma TOF 1.0 (Leco, http://www.leco.com/) and TagFinder 4.0 software on the basis of exact retention times and their corresponding mass spectra (Lüdemann et al. 2008). Full documentation of metabolite profiling data acquisition and interpretation is provided in supplemental dataset (Tables. S1 and S2) online following recommended guidelines (Fernie et al. 2011).
RNA extraction and qRT-PCR analysis of gene expression
Quantitative Real Time PCR was carried out exactly as described in Czechowski et al. (2004). (Czechowski et al. 2004). Primers used here are described in the Table S3. RNA was extracted from at least five biological replicates Extraction of total RNA from leaves, flowers and siliques was performed using TRIzol reagent (Invitrogen); from roots as described in Bugos et al. (1995) with minor modifications; and from seeds as described in Birtic and Kranner (2006). Digestion with DNAse I (Ambion) was performed according to the manufacturer’s instructions. To confirm the absence of genomic DNA contamination, a quantitative PCR analysis using specific primer pairs was performed. The primer pairs were designed to amplify intron sequence of Actin2 (At3g18780, ACT2 forward 5′-ACTTTCATCAGCCGTTTTGA-3′ and reverse 5′-ACGATTGGTTGAATATCATCAG-3′). The integrity of the RNA was checked on 1 % (w/v) agarose gels, and the concentration was measured before and after DNase I digestion using a Nanodrop ND-1000 spectrophotometer. cDNA was synthesized from 2 μg total RNA using Superscript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. The efficiency of cDNA synthesis was estimated by quantitative PCR using two primer pairs amplifying the 5′ and 3′ regions of glyceraldehyde 3-phosphate dehydrogenase (GAPDH5′, forward primer 5′-TCTCGATCTCAATTTCGCAAAA-3′/reverse primer 5′-CGAAACCGTTGATTCCGATTC-3′ and GAPDH3′ forward 5′-TTGGTGACAACAGGTCAAGCA-3′/reverse primer 5′-AAACTTGTCGCTCAATGCAATC-3′). The PCR reaction was performed in a 5 μL volume as described by Caldana et al. (2007). Data analysis was performed using SDS software version 2.4 (Applied Biosystems). Genes expressions were normalized against the constitutively expressed ubiquitin (UQ) using the following primer pair 5´-AGCAGTTGGAGGATGGCAGAAC-3´ and 5´-CGGAGCCTGAGAACAAGATGAAGG-3′.
Statistical analysis
The term significant is used here only when the change in question has been confirmed to be significant (Pp < 0.05 or Pp < 0.01) with the Student’s t test. All statistical analyses were performed using the algorithm embedded into Microsoft Excel.
References
Adams-Phillips L, Briggs AG, Bent AF (2010) Disruption of poly(ADP-ribosyl)ation mechanisms alters responses of Arabidopsis to biotic stress. Plant Physiol 152:267–280. doi:10.1104/pp.109.148049
Ame JC, Spenlehauer C, de Murcia G (2004) The PARP superfamily. Bioessays 26:882–893. doi:10.1002/bies.20085
Amor Y, Babiychuk E, Inze D, Levine A (1998) The involvement of poly(ADP-ribose)polymerase in the oxidative stress responses in plants. FEBS Lett 440:1–7. doi:10.1016/s0014-5793(98)01408-2
Andrabi SA et al (2006) Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci USA 103:18308–18313. doi:10.1073/pnas.0606526103
Ap Rees T, Beevers H (1960) Pentose phosphate pathway as a major component of induced respiration of carrot and potato slices. Plant Physiol 35:839–847
Babiychuk E, Van Montagu M, Kushnir S (2001) N-terminal domains of plant poly(ADP-ribose)polymerases define their association with mitotic chromosomes. Plant J 28:245–255. doi:10.1046/j.1365-313X.2001.01143.x
Bai P et al (2011a) PARP2 Regulates SIRT1 expression and whole-body energy expenditure. Cell Metab 13:450–460. doi:10.1016/j.cmet.2011.03.013
Bai P et al (2011b) PARP1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab 13:461–468. doi:10.1016/j.cmet.2011.03.004
Bauwe H, Hagemann M, Fernie AR (2010) Photorespiration: players, partners and origin. Trends Plant Sci 15:330–336. doi:10.1016/j.tplants.2010.03.006
Belenky P, Bogan KL, Brenner C (2007) NAD+ metabolism in health and disease. Trends Biochem Sci 32:12–19. doi:10.1016/j.tibs.2006.11.006
Berglund T, Ohlsson A (1995) Defensive and secondary metabolism in plant tissue cultures, with special reference to nicotinamide, glutathione and oxidative stress. Plant Cell Tiss Organ Cult 43:137–145. doi:10.1007/bf00052169
Birtic S, Kranner I (2006) Isolation of high-quality RNA from polyphenol-, polysaccharide- and lipid-rich seeds. Phytochem Anal 17:144–148
Boltz KA, Jasti M, Townley JM, Shippen DE (2014) Analysis of poly(ADP-ribose) polymerases in Arabidopsis telomere biology. PLoS One. doi:10.1371/journal.pone.0088872
Briggs AG, Bent AF (2011) Poly(ADP-ribosyl)ation in plants. Trends Plant Sci 16:372–380. doi:10.1016/j.tplants.2011.03.008
Bugos RC, Chiang VL, Zhang XH, Campbell ER, Podila GK, Campbell WH (1995) RNA isolation from plant tissues recalcitrant to extraction in guanidine. Biotechniques 19:734–737
Bürkle A (2001) Physiology and pathophysiology of poly(ADP-ribosyl)ation. BioEssays 23:795–806
Caldana C, Scheible W, Mueller-Roeber B, Ruzicic S (2007) A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3:7. doi:10.1186/1746-4811-3-7
Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38:366–379. doi:10.1111/j.1365-313X.2004.02051.x
de Block M, Verduyn C, De Brouwer D, Cornelissen M (2005) Poly(ADP-ribose)polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J 41:95–106. doi:10.1111/j.1365-313X.2004.02277.x
Essemine J, Govindachary S, Joly D, Ammar S, Bouzid S, Carpentier R (2012) Effect of moderate and high light on photosystem II function in Arabidopsis thaliana depleted in digalactosyl-diacylglycerol. Biochim Biophys Acta 1817:1367–1373. doi:10.1016/j.bbabio.2012.02.004
Fernie AR, Carrari F, Sweetlove LJ (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7:254–261. doi:10.1016/j.pbi.2004.03.007
Fernie AR et al (2011) Recommendations for reporting metabolite data. Plant Cell 23:2477–2482. doi:10.1105/tpc.111.086272
Folta KM, Kaufman LS (2006) Isolation of Arabidopsis nuclei and measurement of gene transcription rates using nuclear run-on assays. Nat Protoc 1:3094–3100. doi:10.1038/nprot.2006.471
Grube K, Bürkle A (1992) Poly(ADP-ribose)polymerase activity in mononuclear leukocytes of 13 mammalian species correlates with species-specific life span. Proc Natl Acad Sci USA 89:11759–11763
Hashida SN, Takahashi H, Uchimiya H (2009) The role of NAD+ biosynthesis in plant development and stress responses. Ann Bot 103:819–824. doi:10.1093/aob/mcp019
Heeres JT, Hergenrother PJ (2007) Poly(ADP-ribose) makes a date with death. Curr Opin Chem Biol 11:644–653. doi:10.1016/j.cbpa.2007.08.038
Hunt L, Gray JE (2009) The relationship between pyridine nucleotides and seed dormancy. New Phytol 181:62–70. doi:10.1111/j.1469-8137.2008.02641.x
Hunt L, Lerner F, Ziegler M (2004) NAD+—new roles in signalling and gene regulation in plants. New Phytol 163:31–44. doi:10.1111/j.1469-8137.2004.01087.x
Hunt L, Holdsworth MJ, Gray JE (2007) Nicotinamidase activity is important for germination. Plant J 51:341–351. doi:10.1111/j.1365-313X.2007.03151.x
Jia Q, Dulk-Ras A, Shen H, Hooykaas PJ, Pater S (2013) Poly(ADP-ribose)polymerases are involved in microhomology mediated back-up non-homologous end joining in Arabidopsis thaliana. Plant Mol Biol 82:339–351. doi:10.1007/s11103-013-0065-9
König A-C et al (2014) The Arabidopsis class II sirtuin is a lysine deacetylase and interacts with mitochondrial energy metabolism. Plant Physiol 164:1401–1414. doi:10.1104/pp.113.232496
Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1:387–396. doi:10.1038/nprot.2006.59
Lüdemann A, Strassburg K, Erban A, Kopka J (2008) TagFinder for the quantitative analysis of gas chromatography-mass spectrometry (GC–MS)-based metabolite profiling experiments. Bioinformatics 24:732–737. doi:10.1093/bioinformatics/btn023
Luo J et al (2001) Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107:137–148
Lytovchenko A, Sweetlove L, Pauly M, Fernie A (2002) The influence of cytosolic phosphoglucomutase on photosynthetic carbohydrate metabolism. Planta 215:1013–1021. doi:10.1007/s00425-002-0826-1
Meyer RG, Meyer-Ficca ML, Jacobson EL, Jacobson MK (2006) Enzymes in poly(ADP-ribose) metabolism. In: Burkle A (ed) Poly(ADP-Ribosyl)ation. Molecular Biology Intelligence Unit, Landes Bioscience, Georgetown, pp 1–12
Nunes-Nesi A et al (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137:611–622. doi:10.1104/pp.104.055566
Nunes-Nesi A et al (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50:1093–1106
Nunes-Nesi A et al (2014) Is there a metabolic requirement for photorespiratory enzyme activities in heterotrophic tissues? Mol Plant 7:248–251. doi:10.1093/mp/sst111
Payne JF, Bal AK (1976) Cytological detection of poly(ADP-ribose)polymerase. Exp Cell Res 99:428–432
Pellny TK, Locato V, Vivancos PD, Markovic J, De Gara L, Pallardó FV, Foyer CH (2009) Pyridine nucleotide cycling and control of intracellular redox state in relation to poly(ADP-ribose)polymerase activity and nuclear localization of glutathione during exponential growth of Arabidopsis cells in culture. Mol Plant 2:442–456. doi:10.1093/mp/ssp008
Pétriacq P et al (2012) Inducible NAD+ overproduction in Arabidopsis alters metabolic pools and gene expression correlated with increased salicylate content and resistance to Pst-AvrRpm1. Plant J 70:650–665. doi:10.1111/j.1365-313X.2012.04920.x
Rissel D, Losch J, Peiter E (2014) The nuclear protein poly(ADP-ribose) polymerase3 (AtPARP3) is required for seed storability in Arabidopsis thaliana. Plant Biol 16:1058–1064. doi:10.1111/plb.12167
Sallmann FR, Vodenicharov MD, Wang ZQ, Poirier GG (2000) Characterization of sPARP1—an alternative product of PARP1 gene with poly(ADP-ribose)polymerase activity independent of DNA strand breaks. J Biol Chem 275:15504–15511
Schippers JHM, Nunes-Nesi A, Apetrei R, Hille J, Fernie AR, Dijkwel PP (2008) The Arabidopsis onset of leaf death5 Mutation of quinolinate synthase affects nicotinamide adenine dinucleotide biosynthesis and causes early ageing. Plant Cell 20:2909–2925. doi:10.1105/tpc.107.056341
Schulz P et al (2012) Chemical PARP inhibition enhances growth of Arabidopsis and reduces anthocyanin accumulation and the activation of stress protective mechanisms. PLoS One. doi:10.1371/journal.pone.0037287
Schulz P et al (2014) Poly(ADP-ribose)polymerase activity controls plant growth by promoting leaf cell number. PLoS One 9:e90322. doi:10.1371/journal.pone.0090322
Sienkiewicz-Porzucek A et al (2010) Mild reductions in mitochondrial NAD+-dependent isocitrate dehydrogenase activity result in altered nitrate assimilation and pigmentation but do not impact growth. Mol Plant 3:156–173. doi:10.1093/mp/ssp101
Song J, Keppler BD, Wise RR, Bent AF (2015) PARP2 is the predominant poly(ADP-ribose) polymerase in Arabidopsis DNA damage and immune reponses. PLoS Genet 11:e1005200
Takahara K, Kasajima I, Takahashi H, Hashida S, Itami T, Onodera H, Toki S, Yanagisawa S, Kawai-Yamada M, Uchimiya H (2010) Metabolome and photochemical analysis of rice plants overexpressing Arabidopsis NAD kinas gene. Plant Physiol 152:1863–1873
Takahashi H, Takahara K, Hashida S, Hirabayashi T, Fujimori T, Kawai-Yamada M, Yamaya T, Yanagisawa S, Uchimiya H (2009) Pleiotropic modulation of carbon and nitrogen metabolism in Arabidopsis plants overexpressing the NAD kinase 2 gene. Plant Physiol 151:100–113
Tian RH, Chen DP, Dai YR (1999) Assaying poly(ADP-ribose)polymerase activity in plants by polarograph method. Chin Sci Bull 44:1883–1887
Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The botany array resource: e-Northerns, expression angling, and promoter analyses. Plant J 43:153–163. doi:10.1111/j.1365-313X.2005.02437.x
Trucco C, Javier Oliver F, de Murcia G, Ménissier-de Murcia J (1998) DNA repair defect in poly(ADP-ribose)polymerase-deficient cell lines. Nucl Acids Res 26:2644–2649. doi:10.1093/nar/26.11.2644
Vanderauwera S, De Block M, van de Steene N, de Cottet BV, Metzlaff M, Van Breusegem F (2007) Silencing of poly(ADP-ribose)polymerase in plants alters abiotic stress signal transduction. Proc Natl Acad Sci USA 104:15150–15155. doi:10.1073/pnas.0706668104
Wahl V et al (2013) Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339:704–707. doi:10.1126/science.1230406
Whitby AJ, Whish WJ (1977) Poly(adenose diphosphate ribose) in wheat. Biochem Soc Trans 5:948–949
Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape-Arabidopsis sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods. doi:10.1186/1746-4811-5-16
Acknowledgments
This work was supported by fundings from the Max Planck Society (P.A.P., V.W., T.T., J.J.O., M.S., W.L.A. and A.R.F.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Pham, P.A., Wahl, V., Tohge, T. et al. Analysis of knockout mutants reveals non-redundant functions of poly(ADP-ribose)polymerase isoforms in Arabidopsis. Plant Mol Biol 89, 319–338 (2015). https://doi.org/10.1007/s11103-015-0363-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-015-0363-5