Abstract
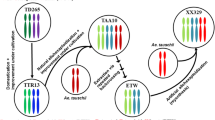
The formation and evolution of common wheat (Triticum aestivum L., genome BBAADD) involves allopolyploidization events at two ploidy levels. Whether the two ploidy levels (tetraploidy and hexaploidy) have impacted the BBAA subgenomes differentially remains largely unknown. We have reported recently that extensive and distinct modifications of transcriptome expression occurred to the BBAA component of common wheat relative to the evolution of gene expression at the tetraploid level in Triticum turgidum. As a step further, here we analyzed the genetic and cytosine DNA methylation differences between an extracted tetraploid wheat (ETW) harboring genome BBAA that is highly similar to the BBAA subgenomes of common wheat, and a set of diverse T. turgidum collections, including both wild and cultivated genotypes. We found that while ETW had no significantly altered karyotype from T. turgidum, it diverged substantially from the later at both the nucleotide sequence level and in DNA methylation based on molecular marker assay of randomly sampled loci across the genome. In particular, ETW is globally less cytosine-methylated than T. turgidum, consistent with earlier observations of a generally higher transcriptome expression level in ETW than in T. turgidum. Together, our results suggest that genome evolution at the allohexaploid level has caused extensive genetic and DNA methylation modifications to the BBAA subgenomes of common wheat, which are distinctive from those accumulated at the tetraploid level in both wild and cultivated T. turgidum genotypes.





Similar content being viewed by others
References
Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8:135–141
Akhunova AR, Matniyazov RT, Liang H, Akhunov ED (2010) Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Gen 11:505
Badaeva ED, Dedkova OS, Gay G, Pukhalskyi VA, Zelenin AV, Bernard S, Bernard M (2007) Chromosomal rearrangements in wheat: their types and distribution. Genome 50:907–926
Buggs RJ, Zhang L, Miles N, Tate JA, Gao L, Wei W, Schnable PS, Barbazuk WB, Soltis PS, Soltis DE (2011) Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr Biol 21:551–556
Chagué V, Just J, Mestiri I, Balzergue S, Tanguy AM, Huneau C, Huteau V, Belcram H, Coriton O, Jahier J (2010) Genome-wide gene expression changes in genetically stable synthetic and natural wheat allohexaploids. New Phytol 187:1181–1194
Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345:950–953
Chen ZJ, Ni Z (2006) Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. BioEssays 28:240–252
Comai L (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6:836–846
Diez CM, Roessler K, Gaut BS (2014) Epigenetics and plant genome evolution. Curr Opin Plant Biol 18:1–8
Dong Z, Wang Y, Zhang Z, Shen Y, Lin X, Ou X, Han F, Liu B (2006) Extent and pattern of DNA methylation alteration in rice lines derived from introgressive hybridization of rice and Zizania latifolia Griseb. Theor Appl Genet 113:196–205
Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF (2008) Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet 42:443–461
Dubcovsky J, Dvorak J (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316:1862–1866
Dvořák J (1976) The relationship between the genome of Triticum urartu and the A and B genomes of Triticum aestivum. Can J Genet Cytol 18:371–377
Dvorak J, Akhunov ED (2005) Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops–Triticum alliance. Genetics 171:323–332
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Feldman M (2000) Origin of cultivated wheat. The world wheat book: a history of wheat breeding
Feldman M, Levy AA (2012) Genome evolution due to allopolyploidization in wheat. Genetics 192:763–774
Feldman M, Liu B, Segal G, Abbo S, Levy AA, Vega JM (1997) Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147:1381–1387
Feldman M, Levy AA, Fahima T, Korol A (2012) Genomic asymmetry in allopolyploid plants: wheat as a model. J Exp Bot 63:5045–5059
Flagel LE, Wendel JF, Udall JA (2012) Duplicate gene evolution, homoeologous recombination, and transcriptome characterization in allopolyploid cotton. BMC Gen 13:302
Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC (2007) Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19:3403–3417
Gornicki P, Zhu H, Wang J, Challa GS, Zhang Z, Gill BS, Li W (2014) The chloroplast view of the evolution of polyploid wheat. New Phytol 204:704–714
Grover C, Gallagher J, Szadkowski E, Yoo M, Flagel L, Wendel J (2012) Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol 196:966–971
Han F, Liu B, Fedak G, Liu Z (2004) Genomic constitution and variation in five partial amphiploids of wheat–Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theor Appl Genet 109:1070–1076
Han F, Fedak G, Guo W, Liu B (2005) Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGc1R-1a) in newly synthesized wheat allopolyploids. Genetics 170:1239–1245
Hegarty MJ, Barker GL, Wilson ID, Abbott RJ, Edwards KJ, Hiscock SJ (2006) Transcriptome shock after interspecific hybridization in senecio is ameliorated by genome duplication. Curr Biol 16:1652–1659
Heslop-Harrison JS, Murata M, Ogura Y, Schwarzacher T, Motoyoshi F (1999) Polymorphisms and genomic organization of repetitive DNA from centromeric regions of Arabidopsis chromosomes. Plant Cell 11:31–42
Hu Z, Han Z, Song N, Chai L, Yao Y, Peng H, Ni Z, Sun Q (2013) Epigenetic modification contributes to the expression divergence of three TaEXPA1 homoeologs in hexaploid wheat (Triticum aestivum). New Phytol 197:1344–1352
Huang S, Sirikhachornkit A, Su X, Faris J, Gill B, Haselkorn R, Gornicki P (2002) Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci USA 99:8133–8138
Jackson S, Chen ZJ (2010) Genomic and expression plasticity of polyploidy. Curr Opin Plant Biol 13:153–159
Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100
Kenan-Eichler M, Leshkowitz D, Tal L, Noor E, Melamed-Bessudo C, Feldman M, Levy AA (2011) Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188:263–272
Kerber E (1964) Wheat: reconstitution of the tetraploid component (AABB) of hexaploids. Science 143:253–255
Kraitshtein Z, Yaakov B, Khasdan V, Kashkush K (2010) Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics 186:801–812
Li A, Liu D, Wu J, Zhao X, Hao M, Geng S, Yan J, Jiang X, Zhang L, Wu J (2014) mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26:1878–1900
Liu B, Vega J, Feldman M (1998a) Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. I. Changes in low-copy noncoding DNA sequences. Genome 41:272–277
Liu B, Vega J, Feldman M (1998b) Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome 41:535–542
Lukens LN, Pires JC, Leon E, Vogelzang R, Oslach L, Osborn T (2006) Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol 140:336–348
Madlung A, Wendel J (2013) Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet Genome Res 140:270–285
Madlung A, Masuelli RW, Watson B, Reynolds SH, Davison J, Comai L (2002) Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol 129:733–746
Matsuoka Y (2011) Evolution of polyploid Triticum wheats under cultivation: the role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol 52:750–764
Mayfield D, Chen ZJ, Pires JC (2011) Epigenetic regulation of flowering time in polyploids. Curr Opin Plant Biol 14:174–178
McClelland M, Nelson M, Raschke E (1994) Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucl Acid Res 22:3640–3659
McClintock B (1984) The significance of responses of the genome to challenge. Science 226:792–801
Olsen KM, Wendel JF (2013) Crop plants as models for understanding plant adaptation and diversification. Front Plant Sci 4: art no 290
Ozkan H, Levy AA, Feldman M (2001) Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops–Triticum) group. Plant Cell 13:1735–1747
Parisod C, Salmon A, Zerjal T, Tenaillon M, Grandbastien MA, Ainouche M (2009) Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol 184:1003–1015
Paun O, Bateman RM, Fay MF, Luna JA, Moat J, Hedrén M, Chase MW (2011) Altered gene expression and ecological divergence in sibling allopolyploids of Dactylorhiza (Orchidaceae). BMC Evol Biol 11: art no 113
Pérez-Figueroa A (2013) msap: a tool for the statistical analysis of methylation-sensitive amplified polymorphism data. Mol Ecol Res 13:522–527
Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR, Mayer KF, Olsen O-A (2014) Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345:1250091
Pires JC, Zhao J, Schranz M, Leon EJ, Quijada PA, Lukens LN, Osborn TC (2004) Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol J Lin Soc 82:675–688
Pont C, Murat F, Guizard S, Flores R, Foucrier S, Bidet Y, Quraishi UM, Alaux M, Doležel J, Fahima T (2013) Wheat syntenome unveils new evidences of contrasted evolutionary plasticity between paleo-and neoduplicated subgenomes. Plant J 76:1030–1044
Pritchard J, Wen W, Falush D (2004) Documentation for Structure software: version 2. Department of Human Genetics, University of Chicago, Chicago
Qi B, Huang W, Zhu B, Zhong X, Guo J, Zhao N, Xu C, Zhang H, Pang J, Han F, Liu B (2012) Global transgenerational gene expression dynamics in two newly synthesized allohexaploid wheat (Triticum aestivum) lines. BMC Biol 10: art no 3
Saintenac C, Jiang D, Akhunov ED (2011) Targeted analysis of nucleotide and copy number variation by exon capture in allotetraploid wheat genome. Genome Biol 12:R88
Salamini F, Özkan H, Brandolini A, Schäfer-Pregl R, Martin W (2002) Genetics and geography of wild cereal domestication in the Near East. Nat Rev Genet 3:429–441
Salmon A, Ainouche ML, Wendel JF (2005) Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol Ecol 14:1163–1175
Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA (2001) Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13:1749–1759
Shitsukawa N, Tahira C, K-i Kassai, Hirabayashi C, Shimizu T, Takumi S, Mochida K, Kawaura K, Ogihara Y, Murai K (2007) Genetic and epigenetic alteration among three homoeologous genes of a class E MADS box gene in hexaploid wheat. Plant Cell 19:1723–1737
Soltis PS, Soltis DE (2009) The role of hybridization in plant speciation. Annu Rev Plant Biol 60:561–588
Testillano PS, Solis MT, Risueno MC (2013) The 5-methyl-deoxy-cytidine (5mdC) localization to reveal in situ the dynamics of DNA methylation chromatin pattern in a variety of plant organ and tissue cells during development. Physiol Plant 149:104–113
The International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345(6194):1251788
Van de Peer Y, Maere S, Meyer A (2009) The evolutionary significance of ancient genome duplications. Nat Rev Genet 10:725–732
Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249
Xu Y, Zhong L, Wu X, Fang X, Wang J (2009) Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta 229:471–483
Yaakov B, Kashkush K (2012) Mobilization of Stowaway-like MITEs in newly formed allohexaploid wheat species. Plant Mol Biol 80:419–427
Yoo MJ, Szadkowski E, Wendel JF (2013) Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110:171–180
Yoo MJ, Liu XX, Pires JC, Soltis PS, Soltis DE (2014) Nonadditive gene expression in polyploids. Annu Rev Genet 48:485–517
Zhang MS, Yan HY, Zhao N, Lin XY, Pang JS, Xu KZ, Liu LX, Liu B (2007) Endosperm-specific hypomethylation, and meiotic inheritance and variation of DNA methylation level and pattern in sorghum (Sorghum bicolor L.) inter-strain hybrids. Theor Appl Genet 115:195–207
Zhang W, Lee H-R, Koo D-H, Jiang J (2008) Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell 20:25–34
Zhang H, Bian Y, Gou X, Zhu B, Xu C, Qi B, Li N, Rustgi S, Zhou H, Han F (2013) Persistent whole-chromosome aneuploidy is generally associated with nascent allohexaploid wheat. Proc Natl Acad Sci USA 110:3447–3452
Zhang H, Zhu B, Qi B, Gou X, Dong Y, Xu C, Zhang B, Huang W, Liu C, Wang X (2014) Evolution of the BBAA component of bread wheat during its history at the allohexaploid level. Plant Cell 26:2761–2776
Zhao N, Zhu B, Li M, Wang L, Xu L, Zhang H, Zheng S, Qi B, Han F, Liu B (2011) Extensive and heritable epigenetic remodeling and genetic stability accompany allohexaploidization of wheat. Genetics 188:499–510
Zohary D, Feldman M (1962) Hybridization between amphidiploids and the evolution of polyploids in the wheat (Aegilops–Triticum) Group. Evolution 16:44–61
Acknowledgments
We thank Prof. Moshe Feldman of the Weizmann Institute of Science, Israel, for providing the initial seeds of ETW and its common wheat donor. We are grateful to an anonymous reviewer for constructive comments to improve the manuscript. This work was supported by the National Natural Science Foundation of China (31290210, 11301064, 31170208) and the Program for Introducing Talents to Universities (B07017).
Conflict of interests
The authors have declared that no competing interests exist.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, C., Yang, X., Zhang, H. et al. Genetic and epigenetic modifications to the BBAA component of common wheat during its evolutionary history at the hexaploid level. Plant Mol Biol 88, 53–64 (2015). https://doi.org/10.1007/s11103-015-0307-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-015-0307-0