Abstract
Codon usage bias is the preferential or non-random use of synonymous codons, a ubiquitous phenomenon observed in bacteria, plants and animals. Different species have consistent and characteristic codon biases. Codon bias varies not only with species, family or group within kingdom, but also between the genes within an organism. Codon usage bias has evolved through mutation, natural selection, and genetic drift in various organisms. Genome composition, GC content, expression level and length of genes, position and context of codons in the genes, recombination rates, mRNA folding, and tRNA abundance and interactions are some factors influencing codon bias. The factors shaping codon bias may also be involved in evolution of the universal genetic code. Codon-usage bias is critical factor determining gene expression and cellular function by influencing diverse processes such as RNA processing, protein translation and protein folding. Codon usage bias reflects the origin, mutation patterns and evolution of the species or genes. Investigations of codon bias patterns in genomes can reveal phylogenetic relationships between organisms, horizontal gene transfers, molecular evolution of genes and identify selective forces that drive their evolution. Most important application of codon bias analysis is in the design of transgenes, to increase gene expression levels through codon optimization, for development of transgenic crops. The review gives an overview of deviations of genetic code, factors influencing codon usage or bias, codon usage bias of nuclear and organellar genes, computational methods to determine codon usage and the significance as well as applications of codon usage analysis in biological research, with emphasis on plants.
Similar content being viewed by others
Introduction
The genetic language of DNA base sequences is deciphered as twenty-letter language of amino acids in proteins, in multiple ways. Out of the 64 nucleotide triplets (codons), 61 encode standard 20 amino acids, whereas three are translation stop signals. Degeneracy of genetic code permits the same amino acid to be encoded by different codons or synonymous codons. Tryptophan and methionine are encoded by a single codon, while remaining 18 of 20 amino acids are encoded by multiple synonymous codons [1]. However, synonymous codons are not randomly or equally used, but some repeatedly preferred over others, to code for an amino acid. This universal phenomenon of usage of synonymous codons with different frequencies is termed as codon usage bias (CUB). CUB is widespread across species and serves as a code within the genetic code or the second genetic code [2, 3]. Biased frequency of synonymous codons (CUB) varies not only among genomes, but also among functionally related genes and within a single gene [4]. Reasons for existence of codon usage bias in organisms are intriguing. Mutations in gene-coding regions, especially in the second or third nucleotide of an existing codon, that exchange one synonymous codon for another, do not change the amino acid specified by new, modified codons nor the peptide primary sequence. Such synonymous or ‘silent’ mutations without any functional consequences, when selected during evolution, cause synonymous codon usage biases in genomes [5]. Thus, codon usage bias exists due to biased mutational patterns, whereby some codons may be more prone to mutation than others and maintained by selection [6]. GC heterogeneity and GC-biased gene conversion (gBGC) also shape codon usage bias in a manner dependent upon the local recombination rate. Evolution of synonymous codon usage is therefore a result of balance between mutation, natural selection and genetic drift on translational efficiency of genes and may contribute to genome evolution in a significant manner [7,8,9]. Mutational mechanism of codon bias postulates that codon bias is due to biases in nucleotides produced by point mutations, or in the rates or repair of point mutations and explains for the interspecific variation in codon usage. Meanwhile, theory of natural selection postulates that synonymous mutations influencing fitness of an organism are promoted or repressed throughout evolution, resulting in codon usage variations across a genome or a gene [10].
Codon bias plays an important role in a multitude of cellular processes, such as transcription, mRNA stability, translation efficiency and accuracy, as well as structure, expression, function and co-translational folding of proteins [1, 3, 6]. Codon bias determines transcription levels by affecting chromatin structures and mRNA folding and affects translation efficiency, by tuning the elongation rate of translation, suggesting that codon bias is due to genome adaptation to both transcription and translation machineries [1, 11]. Selection exercised on gene sequences without amino acid changes has profound implications in the study of molecular evolution of genes. Codon bias analysis can reveal horizontal gene transfers and evolutionary relationships between organisms, since closely related organisms have similar patterns of codon usage [6]. Highly expressed proteins are mostly encoded by genes with mostly optimal codons [12]. Hence, the most important application of CUB is in genetic engineering or recombinant DNA technology, where codon optimization of genes from alien sources are carried out to enhance protein expression for heterologous genes [13].
Striding into the post genomic era, marked by an explosion of information on genetic sequences, it becomes imperative that the databases generated are analysed to extract meaningful information. Many computational approaches are already devised to analyze sequence similarities [14]. However, research on codon usage bias (CUB) is under-exploited at the genomic level. Plants exhibit variations in gene expression, physiology and stress response under diverse environmental conditions. Knowledge of codon usage of plants will therefore help in understanding the molecular mechanisms of environmental adaptation and biological diversity of each plant species [9]. The review discusses of the fundamentals of genetic code and its deviations, factors affecting codon usage bias, CUB in plants with reference to the nuclear as well as chloroplast genes, computational methods to determine CUB and applications of codon usage in crop improvement.
Degeneracy and deviances of genetic code
Deciphering the enigma termed genetic code and addressing the coding problem of how the 4 nucleotides in the DNA were translated to the 20 amino acids in proteins, was the biggest challenge after the discovery of DNA structure and formulation of Central Dogma of molecular biology in 1953. George Gamow’s hypothetical “Diamond code”, of overlapping triplet code in 1953, Marshal Nirenberg and Heinrich Matthaei’s experiments on cell-free protein synthesis systems to synthesise poly phenyl alanine and poly proline using polyU and polyA mRNA in 1960, and the experiments using acridine-induced single base mutations in DNA of rII cistronic region of T4 phage in 1961 by Francis Crick and colleagues, proved that genetic code is triplet, degenerate, non-overlapping and unpunctuated and each nucleotide sequence is read from a specific starting point [15,16,17]. The landmark paper of Francis Crick, Leslie Barnett, Sydney Brenner and Richard Watts-Tobin titled “General nature of the genetic code for proteins” was published in journal ‘Nature’ in 1961 [15]. Har Gobind Khorana decoded the entire genetic code using short defined sequences of DNA and artificial mRNA molecules [18]. Synonymous codons specify the same amino acid, and a codon family includes collection of such synonymous codons, whose maximum size is six and minimum size is one. The universal genetic code and the degeneracy of genetic code are highlighted in Tables 1 and 2.
Origin of genetic code is as mysterious and enigmatic as origin of life. Spontaneous interaction of cosmic and terrestrial chemicals or biomolecules in steaming hydrothermal environments, marked the beginning of life. Peptides were easy to synthesize than RNAs in the primordial environment. As many as 70 naturally occurring amino acids in the prebiotic soup probably originated from carbonaceous chondrites. More than 40 different amino acids were produced in Stanley Miller’s atmospheric spark discharge experiments. Subsequent studies showed that 10 of the 20 naturally occurring amino acids could be generated abiotically under simulated primordial earth conditions [19].
Genetic code formed, as dictated by thermodynamic principles, followed a logical sequence of events, specified as few as 4 amino acids with a simple nucleotide code, in the primordial soup (Fig. 1a and b). The earliest amino acids such as glycine, alanine and glutamic acid had simple structure and could be formed in a variety of environments spontaneously, from purely chemical means, without assistance of protein molecules [20]. In the primordial code, when second base in a codon (P2) is G, Gly and Ser are encoded and when C is in P2, Ala, Ser, Thr and Pro are encoded. The most abundant primordial aminoacids are Gly (with G in P2) and Ala (with C in P2). If G is in P1, regardless of which base is at P2, Gly, Glu or Asp, Ala, and Val are encoded. Thus, a primitive code with G in P1 gave four original codons coding for the four or five most prevalent amino acids in the prebiotic soup., GGN encoding Gly, GCN (Ala), GAN (Glu or Asp), and GUN (Val) (where N is any base). The primordial code specified three types of four L-amino acids viz., two semi-polar (Gly, Ala), one hydrophilic (Asp) and one hydrophobic (Val) amino acid. Later, six more amino acids were recruited from the prebiotic environment for tRNA-mRNA-aaRS (aminoacyl-tRNA synthetase) interactions. The relative abundances of these ten amino acids in the order Gly > Ala > Asp > Glu > Val > Ser > Ile > Leu > Pro > Thr, correlated with the free energies of their synthesis, suggesting that thermodynamics determined their relative amounts. These were precursors for the formation of other ten amino acids along prebiotic pathways. Tryptophan has a complex structure, is comparatively rare in the protein code and hence is one of the latest additions to the code. Due to evolving anabolic pathways, when additional amino acids became available, genetic code expanded stepwise, with increasing number of codons to specify correspondingly increased number of amino acids and to eventually include all 20 common protein amino acids [20]. The complexity grew over time, so that codons were reassigned later to a related amino acid, to minimize the consequences of mutations and translational errors. The current code would thus be a relic of the early code.
Complex cellular components originated by encapsulation of RNA, aminoacids and peptide molecules from prebiotic soup by lipid membranes, to initiate a molecular symbiosis inside the protocells. The prebiotic information system was created step-by-step by these biomolecules, at first plasma membranes, followed by RNA and peptides, then tRNAs, mRNAs, and then ribosomes, for programmed protein synthesis. The demand for a wide range of protein enzymes over peptides, was the main selective pressure for the origin of information-directed protein synthesis, a unique signature of life. Transition from peptide to protein in the peptide or RNA world and the interactions between diverse RNA molecules, amino acids or peptides and various enzymes such as ribozymes, aminoacyl tRNA synthetases etc., led to the gradual evolution of the translation system and the genetic code (Fig. 1a and b) [19].
The code was thought to be invariable in all organisms (‘frozen accident’) [21]. Deviations from the standard genetic code and subtle variations in the codon assignment were found in Archaea, Bacteria, eukaryotic nuclear genomes and organellar genomes, mostly in mitochondrial genome with more than 20 alternative codes [22]. Violations of the universal code are rarer for nuclear genes, the deviations mostly restricted to stop-to-sense reassignments of termination codons, while mitochondrial genetic code deviations included sense-to-sense or even sense-to-stop codon reassignments [21, 23]. Reassignment of one or two termination codons as sense ones is reported in eukaryotes. However, a new variant of the nuclear genetic code with all three standard termination codons reassigned to code for amino acids, was discovered in a clade of trypanosomatids or protists, where UGA was reassigned to encode tryptophan, and UAG as well as UAA (UAR) were reassigned to encode glutamate. Use of both UAG and UAA codons in the coding sequences for glutamate can be explained by G-to-U transversion of the first nucleotide in GAG and GAA codons (for glutamate), respectively. Efficient use of UAG, UAA, or UGA as sense codons not only requires sufficient abundance of cognate aminoacyl-tRNAs, but also specific modifications in the eukaryotic release factor 1(eRF1) to recognise all termination codons, so that the specificity of eRF1 is reduced. However, as termination codon, UAA predominated, UAG was rarely used and UGA was not used [24]. Post-transcriptional base modifications at tRNA anticodons modify the codon or anticodon base-pairing rules (‘wobble rule’). Mutations of the tRNA identity elements also result in codon reassignments. As more organisms were studied, genetic code was found to be no longer universal and frozen, but malleable, continuously evolving and ‘quasi’ universal, due to the widespread deviant codes [22]. The deviations of genetic code and the alternative codons used are given in Table 3 [21, 24,25,26,27,28,29]. A list of alternative translation tables is maintained by NCBI (https://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi) [23].
Genetic code has expanded slowly and may continue to evolve with the evolution of organisms and changing environments. The code structure co-evolved with amino acid biosynthesis pathways (co-evolution theory); or for minimizing the adverse effect of point mutations and translation errors (error-minimization theory) or due to physico-chemical affinity between amino acids and the cognate codons (anticodons) (stereochemical theory) [22]. Code evolution takes place due to recruitment of non-standard or non-canonical amino acids (NSAAs or ncAAs) to non-sense codons (genetic code expansion) or to multiple sense codons (genetic code reprogramming). Selenocysteine (Sec, 21st amino acid) and pyrrolysine (Pyl, 22nd amino acid) can be inserted against non-sense codons UGA and UAG respectively [30]. Thus, 22 amino acids are used in the native translation system. Selenocysteine (Sec) is similar to cysteine and serine, but contains a selenium atom in place of sulfur in cysteine, and oxygen in serine, and is synthesized on its cognate tRNA. Mechanisms of incorporation of non-standard aminoacids are given in Fig. 2a. The precursor of Sec is serine, which attaches to tRNASec by seryl-tRNA synthetase (SerRS), to form Ser-tRNASec. In bacteria, Ser-tRNASec is then converted to Sec-tRNASec by selenocysteine synthase (SelA) in the presence selenophosphate, a selenium donor. In Archaea and eukaryotes, an additional step of phosphorylation of Ser-tRNASec by O-phosphoseryl-tRNA kinase (PSTK) occurs, that results in formation of O-phosphoseryl-tRNASec (Sep-tRNASec). Sec-tRNASec is formed from Sep-tRNASec and selenophosphate, by Sep-tRNA:Sec-tRNA synthase (SepSecS). Sec incorporation at the UGA codon requires the presence of a stem-loop structure, a Sec insertion sequence (SECIS), SECIS binding protein 2 (SBP2), Sec specific elongation factor (EFSec), Sec tRNA, phosphoseryl-tRNA kinase (PSTK), SECp43 and Sec synthase [30, 31]. Selenoproteins are found in animals but are absent in fungi or higher plants, and hence tRNASec is absent in higher plants [32]. However, pyrrolysine is not made on tRNAPyl, but as a free amino acid, which is directly ligated to its cognate tRNA. This is recognized by the standard elongation factor EF-Tu and pyrrolysyl-tRNA synthetase (PylRS), that ligates pyrrolysine to UAG codon [33]. Meanwhile, AUG is standard start codon in all organisms that codes for two amino acids, α-N-formylmethionine (fMet) and methionine (Met), respectively for initiation and elongation, where fMet is used for initiation only in bacteria, mitochondria and chloroplasts and Met is used for initiation in eukaryotes. The pre-translationally formed fMet residue is however co-translationally deformylated by a ribosome-associated deformylase and removed [34]. Initiation at near-cognate or alternative start codons are also found in prokaryotes and eukaryotes, though their occurrence is rare in eukaryotes. Mitochondrial genomes in eukaryotes (humans) use AUA and AUU, and those of prokaryotes use GUG and UUG, as alternate start codons. In prokaryotes, abundant initiation occurs at GUG (12%), UUG (8%), CUG, AUU and occasionally at AUC codons. In prokaryotes, ‘class I’ start codons UUG and GUG can initiate with 12–15% efficiency, while ‘class IIA’ start codons CUG, AUU, AUC, AUA and ACG initiate translation with 1–3% efficiency when compared to AUG. CUG is used as start codon to code for leucine. The 30S preinitiation complex in prokaryotes positions the start codon in the P-site, base-paired with the anticodon of initiator fMet-tRNA. The near-cognate-initiator tRNA codon-anticodon mismatch occurs through a wobble mechanism. The relative efficiencies of near-cognate start codon utilization compared to AUG, are primarily controlled by bacterial initiation factor 3 (IF3). In eukaryotes, scanning model of translation initiation is proposed, where the 43S preinitiation complex with small ribosomal subunit, multiple initiation factors, and initiator tRNA recognizes 7-methylguanosine cap of mRNA and scans 5′-to-3′ till first start codon is recognized. At non-AUG start codons, methionine is incorporated via scanning initiation, suggesting wobble base-pairing between the initiator tRNA and near-cognate codons. The presence of a Kozak consensus sequence (especially an A or G at the –3 position) strongly affects the efficiency of initiation at non-AUG start codons. Exceptions to the scanning initiation mechanism at non-AUG start codons include initiation at CUG codons using leucyl-tRNA and eIF2A (instead of eIF2) and RAN (repeat-associated non-AUG) translation via cap-dependent scanning mechanisms in multiple reading frames [35].Codons in any position within the open reading frame can have dual function as with selenocysteine and pyrrolysine, depending on the availability of a specific type of RNA stem-loop structure in the 3′-untranslated regions. Thus, duality of codon function provides additional recoding events and novel features responsible for evolution of genetic code [30].
Mechanism of incorporation of non-standard amino acids and ribosomal frameshifting. a Incorporation of selenocysteine. In prokaryotes, serine attaches to tRNAsec to form Ser-tRNAsec and then to Sec-tRNAsec. In Archaea and eukaryotes, Ser-tRNAsec is phosphorylated to Sep-tRNAsec and then converted to Sec-tRNAsec. SerRS indicates seryl-tRNA synthetase, SelA:selenocysteine synthase, SelenoP:Seleno phosphate, PSTK: O-phosphoseryl-tRNA kinase and SepSecS: Sep-tRNA:Sec-tRNA synthase. Sec incorporation at UGA codon requires mRNA stem loop structure, Sec insertion sequence (SECIS), SECIS binding protein 2 (SBP2), and Sec specific elongation factor (EFSec). Mechanism of incorporation of selenocysteine in Archae and prokaryotes modified and redrawn from [30]. b Incorporation of non-standard amino acids (NSAAs) in proteins through orthogonal translation system (OTS). Orthogonal tRNA (o-tRNA)/aminoacyl-tRNA synthetase (o-aaRS) pairs from phylogenetically distant organisms are used to charge tRNA with NSAAs. c Programmed -1 ribosomal frameshifting. Ribosome shifts 1 nucleotide towards the 5′ end mRNA. This requires heptanucleotide slippery sequence with consensus of X_XXY_YYZ where X = any nucleotide, Y = A/U, Z = A/C/U, a spacer region, and a 3′- RNA secondary structure
Nearly 51 or more non-standard amino acids have been reported [36]. Proteinogenic non-standard amino acids are formed by post-translational modifications, while non-proteinogenic non-standard amino acids (NPAAs) may arise as metabolic intermediates or isomers of standard amino acids such as ornithine, citrulline, norvaline, norleucine, alloisoleucine etc. More than 250 non-protein amino acids are found in plants of Leguminosae, Sapindaceae, Aceraceae, Hippocastenaceae, Cucurbitaceae etc., especially in seeds of legumes and have defense-related functions such as antiherbivory, antimicrobial or as toxins against invertebrates and vertebrates, or allelochemicals, or in protection against stress, in signaling and nitrogen storage [37]. To incorporate NSAAs site-specifically into proteins for novel properties and diverse applications, an orthogonal translation system (OTS), with orthogonal tRNA (o-tRNA)/aminoacyl-tRNA synthetase (o-aaRS) pairs from phylogenetically distant organisms was used (Fig. 2b). The OTS system does not cross-react with the endogenous amino acids, aaRSs, or tRNAs of the host cell [38].
Codon arrangements and amino acid assignments in the genetic code were non-random. Codons with T or U in the second position encoded hydrophobic amino acids, while those with C or G, and those with A in position 2, respectively encoded semipolar and strongly hydrophilic aminoacids. UGA can code for amino acids: l-selenocysteine, l-tryptophan and glycine [20, 22]. The number of synonymous codons for an amino acid was negatively correlated with its molecular weight, but was positively correlated with its frequency in proteins [22].
Codon overlapping is another deviation from genetic code. Codon overlapping occurs in co-translational frameshifts or recoding and by passing, when the ribosome pauses at a rare codon or mRNA secondary structure, shifts forward or backward by a single nucleotide or two nucleotides, and continues translation in a different reading frame [39]. Meanwhile, RNA polymerase slippage results in transcriptional frame shift. Ribosomal frame shifts can be either − 1 or − 2 (1 or 2 nucleotides towards the 5′ end of mRNA) or + 1 (1 nucleotide towards the 3′ end of mRNA), majority of ribosomal frameshifting being − 1 frame shifts. Programmed − 1 ribosomal frameshifting (-1PRF) is a gene expression mechanism which requires a slippery sequence, a spacer region, and a 3′-adjacent stimulatory RNA secondary structure (stem-loop or pseudoknot). In eukaryotes, the slippery sequence has a consensus heptanucleotide motif X_XXY_YYZ, where X is any three identical nucleotides, Y represents A or U and Z represents A, C or U (Fig. 2c). The slippery sequence may vary in length or content in other organisms. In the tandem slippage model of frameshifting, the ribosomal P-site tRNA anticodon, re-pairs from XXY to overlapping codon XXX, and the A-site anticodon re-pairs from YYZ to YYY. However, frameshifting involving re-pairing at a new, non-overlapping frame codon (hopping/bypassing), is less frequent. Coronaviruses utilize high level − 1 frameshifting for synthesis of their polymerase. In + 1 ribosomal frameshifting, ribosome pauses at a codon sequence encoding a rare amino acid or when amount of tRNA of that codon in the cytosol is low. The ribosome and its associated tRNA slips into the new frame (single tRNA slip rather than two). Frameshifting is therefore dependent on codon combinations and the physiological state of the cell [39, 40]. Alternate initiation codons such as non-AUG triplets are used for proteins of low expression, different sub cellular localisations or distinct biological functions in plants [41]. The ambiguities of genetic code in these cases are but tolerated without loss of fidelity.
Expansion of genetic code is a prospective area of research but still in its infancy. Reassignment of stop and start codons and rare sense codons to new amino acid, artificial synthesis of synthetic nucleotides (XNAs) and novel codons, use of four-base codon strategy for synthesis of peptides with multiple non-natural amino acids and natural promiscuous activity of the aminoacyl-synthase enzyme, offer wide possibilities of expanding the standard genetic code [42, 43].
Codon usage bias
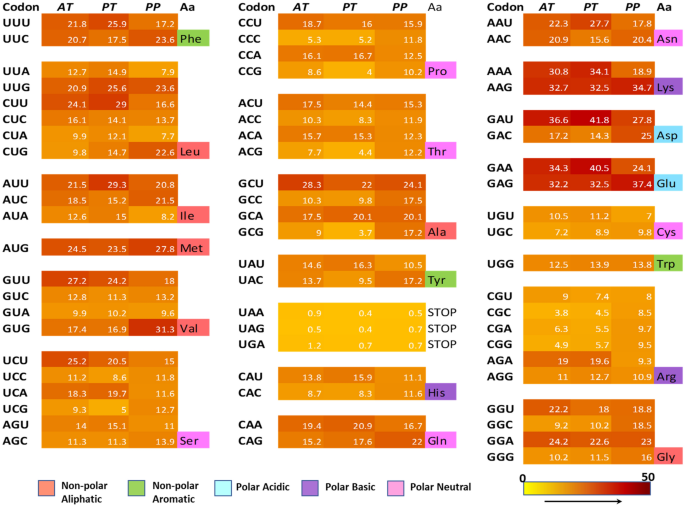
Codon usage bias (CUB) refers to non-uniform use of synonymous codons, the extent of which varies within and among species. Arabidopsis, Populus and Physcomitrella patens are used as model species to analyze codon usage in plants. A comparison of codon usage bias and list of the preferred codons in the three model species are given in Fig. 3 and Table 4 respectively.
Comparison of codon usage bias in model plant species using heat map of codon usage. AT refers to Arabidopsis thaliana, PT: Populus trichocarpa and PP: Physcomitrella patens. The RSCU values for each codon obtained from Kazusa codon database (https://www.kazusa.or.jp/codon/) are indicated inside the box in white. Colour code from 0 to 50 indicates the RSCU values. The amino acids are colour-coded to indicate the groups as shown below. (Color figure online)
Factors affecting codon usage bias
Codon bias or preference varies not only with species of organisms, family or group within kingdom, but also between the genes and between different sites or positions in a gene. Species-specific codon bias is correlated with overall GC content of a genome. Within the organism, among the genes, codon bias may be influenced by expression level and length of gene(s), mRNA structure, amino acid composition and hydrophobicity of the encoded protein and different steps of protein expression [44]. Codon usage is thus dependent on several factors which are summarized in Fig. 4 and detailed below.
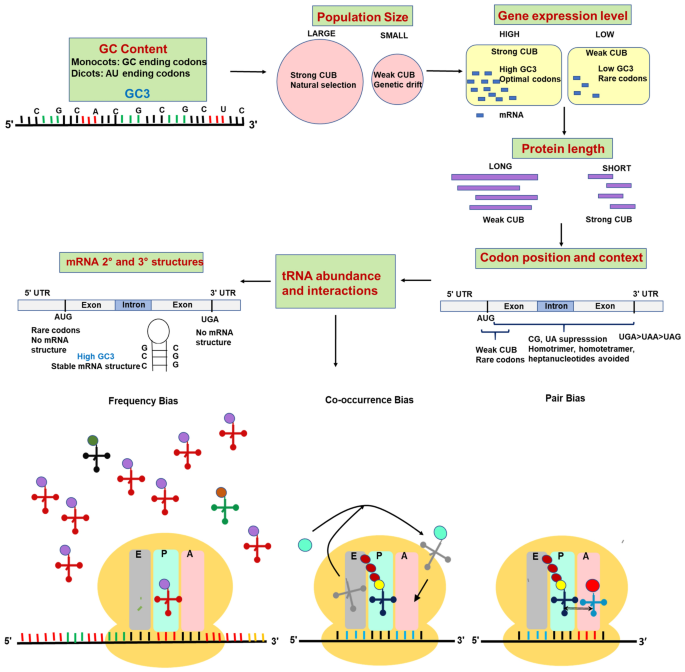
Major factors affecting codon usage bias in organisms such as GC content of genome, population size, gene expression level, protein length, codon position and context, tRNA abundance and interactions and mRNA structure are diagrammatically indicated. tRNA interactions are classified into frequency bias, co-occurence bias and pair bias. E, P and A indicate exit, peptide and amino acid sites in the ribosomes. The tRNA interactions were modified and redrawn from [1]
Factors affecting codon usage bias.
Genome composition
Genomic GC content determined by mutational processes is a major determinant of codon-usage variation across species [10]. Different species have consistent and characteristic codon biases. Plants have a higher G + C content and among plants, monocots have high GC content (59–61%) than dicots (35–42%) [45]. Chloroplast DNA usually possesses a lower GC content (28.5–42%) and plant mitochondrial DNA is comparably GC-richer (40.6–49%). Nuclear gene-coding regions of monocots are enriched in codons ending in C and G, whereas those of dicots have a higher proportion of codons ending in A and U [46]. A-and U-ending codons were predominant in seven different Citrus species and in FAD7 gene of dicots, whereas G- or C-ending codons were used for FAD7 of monocots [2, 47].
In a genome, differences in codon bias between genes may be due to the variation of G + C or dispersion of large isochores homogenous for G + C content, throughout the genome. Composition bias and selection affects codon bias. To correct for intragenomic compositional heterogeneity, direction and strength of the codon bias should be investigated in the local genomic context [48]. Local GC content increases rapidly in recombination hotspots in genome. GC-biased gene conversion depends on local recombination rate and favors fixation of G and C alleles over A and T alleles at polymorphic sites. Overall genome compositional bias affects degenerate 3rd nucleotide position bias of coding sequences. Greater GC content at synonymous positions in the coding regions compared with the flanking introns indicates selection at synonymous sites [49]. In non-grass monocots, a positive relationship was observed between coding and genomic GC content as well as between CAI (Codon adaptation index) and GC3 (GC content in third codon position) [9]. GC3 and codon usage were strongly correlated among genes in rice and Arabidopsis, which suggested that codon usage variations may be due to a mutational bias at the DNA level, rather than natural selection at the translation level [50].
Degree of gene expression
Codon usage may reflect degree of gene expression, though gene expression is determined mainly by transcriptional and post-transcriptional regulations. Codon usage reflects the pool of tRNAs available when a gene is expressed, which in turn depends on the physiological state of the cell [51]. Highly expressed genes have a much stronger codon usage bias and are encoded by optimal codons corresponding to abundant tRNAs, for more rapid and efficient translation. [12, 52]. Codon bias is significantly correlated to gene expression, but independent of base composition. Translational selection acts on highly expressed gene, to have fitness advantage through increased translation efficiency or accuracy [7]. But initiation is rate limiting for production of endogenous proteins and hence the elongation rate should not influence the amount of protein produced [10]. Codon usage and GC content influence mRNA levels by regulating transcription at the chromatin structure level, or through regulation of premature transcription termination and splicing. Thus, codon usage is one of the several factors that affects gene expression through multiple mechanisms at translational, transcriptional and post-transcriptional levels [3].
Only a subset of potential codons is used in highly expressed genes [53]. High G + C content was observed at the silent third nucleotide position in codons of high expression genes [54]. All the preferred codons need not be GC rich and GC/GC3 may not be the accurate representation of codon usage trend [53]. Optimal third (wobble) codon position could end in A or T rather than G or C as frequent alleles [55]. More random and suboptimal codon usage was observed among low expression genes. For genes whose expression varies from lower to higher expression, with a change in environmental conditions, the codon bias is similar to that of highly expressed genes. In Arabidopsis, codon bias was related to the composition of 3′ flanking region, in both strongly and weakly expressed genes [56]. A measure termed as ARSCU (Average ratio of Relative Synonymous Codon Usage of GC-end codons to AT-end codons in each gene) was devised to separate high-expression from low-expression genes, where genes with ARSCU values above threshold value 13 were classified highly expressed under drought [51]. The codon usage bias of highly expressed genes and comparison with lowly expressed genes in few plants are presented in Table 5 [51, 53, 57, 58]. Expression levels of genes could be regulated by virtue of codon strategies.
Pattern of gene expression
Codon bias also depends on whether the genes are spatially or temporally regulated or constitutively and highly expressed. In Arabidopsis, genes encoding abundant protein in vegetal cells such as photosynthetic and housekeeping genes had a highly G/C biased codon usage and genes with strong tissue-specific expression had a weaker A/T-biased codon usage when compared to stress-regulated genes [59]. A unimodal correlation existed between the codon usage bias (CUB) and salt stress-resistant genes among plant species [52]. Tissue-specific genes exhibit significantly different synonymous codon usage in rice, due to GC content variation among tissues, although this effect is weak [8]. Environmental stress factors also may modify codon usage. In a basidiomycete, metabolic specialization to use wood as the sole carbon source had modified codon usage of the genes involved in lignocellulose degradation, to the tRNA pool available, to improve the translation efficiency [60]. A score of modified relative codon bias (MRCBS) was used to elucidate pattern and level of expression of genes in Arabidopsis where, MRCBS and CAI (codon adaptation index) were strongly correlated [53].
Gene length
For same expression pattern, a strong negative correlation exists between frequency of optimal codons, codon usage and gene or protein length. Codon usage decreases with increasing protein length. Codon bias is higher for shorter proteins and lower in longer proteins in yeast, plants and metazoans. However, in E. coli and Saccharomyces cerevisiae, codon-usage bias increased with protein length [4, 50, 61]. In rice genes, weakly expressed genes were longer and had a lower G + C content at the third codon position than strongly expressed genes. Longer transcripts, when enriched in optimal codons ending in G/C, are more likely to form strongly packed mRNA tertiary structures, so that such codons may be counter-selected in genes expressed at low or moderate levels. In shorter genes expressed at high levels, the expression level and frequency of optimal codons ending in G/C are positively correlated [48]. Effect of protein length on codon bias was explained by selection for translation rate [62]. But none of the models could correctly explain the decrease in codon bias for longer proteins.
Codon position and context in the gene
Translation efficiency is determined by both translation initiation and elongation rates, but translation initiation is a predominant factor. Locally biased distributions of rare and frequent codons or “codon landscape” exist in a gene, which may result in variable translation rates [1]. Codon-tRNA co-evolution explains for the bias towards frequent (preferred) codons, which are selected for accurate and efficient translation, while the rare (un-preferred) codons are important in the fitness of the organism, regardless of their position in the coding regions [63]. But un-preferred codons were position-dependent with important functional roles [10]. Synonymous mutations at specific sites may be subject to selection. At the start of a gene, where missense or nonsense errors are less costly, selection is weaker for translation accuracy, but is strong for increasing ribosomal initiation rate and reducing the elongation rate [10]. Rate of synonymous substitutions and SNP density are reduced in 5′ regions, near splicing control elements since synonymous mutations near intron–exon boundaries can create spurious splice sites or disrupt splicing control elements. Also, at 5' end of genes, selection acts for mRNA structure that results in increased usage of A/T rich codons shortly after the gene start. The mRNA secondary structure within the first 40 nucleotides near the 5′ end of a coding region can inhibit ribosomal initiation and hence codons that create strong 5′ mRNA secondary structure are selected against, and one or more rare codons that disrupt 5' mRNA secondary structure are placed at 5′ end of genes in prokaryotes [64]. In E. coli, rare codons are only selected if they are AU-rich, whereas GC-rich codons are repressed. Thus, rare codons are not selected because they are rare, but are selected to weaken or suppress the mRNA structure [65]. Beginning of coding sequences is a region of slow translation. In eukaryotes, reduced codon adaptation in the 5′ region of genes slows down elongation rate thereby reducing the frequency of ribosomal traffic jams towards the 3′ end, and keeps the ribosomes evenly spaced to avoid spontaneous or collision-induced abortions [1]. Also, slow elongation may facilitate recruitment of chaperone proteins to the emergent peptide or in secretion or in membrane localization of nascent protein chain bearing N-terminal signal sequences [10, 64]. Stretches of rare codons cause ribosome pausing or frameshifting, co-translational cleavage of mRNA or amino acid misincorporation and reduces protein yields by obstructing translation initiation [10]. Poorly adapted codons may be selected in sites that require ribosomal pausing, mRNA folding, proper nucleosome positioning, proper co-translational protein folding, ubiquitin modification or in secreted proteins for promoting membrane targeting and secretion efficiency and in highly expressed genes for sequential folding of protein domains during translation [3]. Rare codons are not randomly scattered across genes, but often occur in large clusters, in numerous eukaryotic and prokaryotic genomes and are not always associated with low translation rates. Transcripts enriched in rare codons underwent a higher translation boost than transcripts with common codons [66].
Decreased codon adaptation over the 1st 10–20 codons at 5' end of genes, was observed in plastid and E. coli genes with high overall codon adaptation, but not in those with low codon adaptation [67]. Both lowly and highly expressed genes are similar in their codon usage patterns in the 5′-gene regions, but for highly expressed genes, codon preferences diverge at distal sites resulting in greater positional dependency. Despite the general G + C enrichment by TAMB (Translocation and Assembly Module B) DNA repair system, four-fold enrichment of degenerate codons ending in T was observed in Arabidopsis and rice intergenic DNA, but no such differences were observed in introns [48].
Specific, preferred or bias against certain sequences or avoided nucleotide patterns were observed in the coding region, which differed among species. In prokaryotes, the sequences GAGG and GGAG in the Shine-Dalgarno sequence (UAA GGA GG) were rarely used in the coding region to avoid internal translation sites [67]. Although G/C ending codons were enriched in monocots, the frequency of GGG and CCC codons were not increased, to prevent mRNA tertiary structures. Similarly, CpG dinucleotide and codon GTA were suppressed, to discourage deleterious methylation/deamination events and insertion events that target the CpG and TpA dinucleotides, respectively [48, 69]. NCG codons are avoided in species with a high level of DNA methylation, to avoid mutations because, methylated cytosine (C) in CG dinucleotide, when unrepaired, is easily deaminated into thymine (T), resulting in the conversion of CpG to TpG and the G in the 3rd codon position is wobbly [70]. CpG suppression is observed in coding regions of plants, but not in animal mitochondria or chloroplast genomes which lack methylase activity. Frequency reduction of CpG dinucleotide that exhibits greatest thermodynamic stacking energy of all dinucleoides, might be to facilitate DNA replication and transcription. When G + C level is increased, CpG shortage is decreased [70]. NCG:NCC ratio index which shows methylation level in mRNA coding sequences, is widely used to estimate CpG suppression. Species with a high methylation level such as Populus have a relatively lower NCG:NCC ratio (0.46), while species with a low methylation level such as Arabidopsis have a relatively higher value (0.921). NUA codons also had low codon usage since UA dinucleotides are sites for RNA hydrolysis by ribonucleases. Therefore, UpA suppression reduces mRNA degradation and increases protein production [71]. Heptanucleotides that are prone to frameshifts and codons promoting formation of complex mRNA tertiary structures, are under-represented in the coding sequences [69]. Codon frequencies are modified to avoid homotrimer and homotetramer formation especially for G and C than A and T [70]. G and C homotetramers were avoided in Graminae whereas T and A tetranucleotides were omitted in dicots. Intercodon CpGs and TpAs were preferably replaced by TpGs and CpAs, respectively. Two out of three stop codons start with TpA. TpA is energetically less stable than all other dinucleotides, which provides flexibility for untwisting and bending of DNA double helix and hence are found in TATA sequences and at replication origins. Reduction of TpA therefore avoids inappropriate binding of regulatory factors [70].
Non-random distribution or clustering of iso-accepting codons (synonymous codons decoded by the same anticodon of tRNA) in the coding region and genome is termed as codon co-occurrence [72]. Codon context meanwhile refers to the nucleotides or codons adjacent to a codon. Compositional context can also influence synonymous codon selection, and this phenomenon is known as context-dependent codon bias [48]. Codon context bias shows preferences for a codon pair within an organism, that can have conserved patterns and may be species-specific. Codon context bias can influence missense and nonsense suppression, elongation rates and translational accuracy, since different species have varied abundance of tRNA iso-acceptors for each codon family [69, 73]. In many eukaryotic species, both synonymous and non-synonymous mutations are selected to maintain context biases [49]. Codon-pair context is mainly determined by constraints imposed by the translational machinery in eubacteria and archeae, and by DNA methylation and trinucleotide repeats in eukaryotes. Context preferences exist in coding as well as non-coding sequences. Lysine is preferentially encoded by AAA, if guanosine is 3' adjacent (AAAG), but by AAG, if cytidine is 3' adjacent (AAGC). A less significant bias is observed at the 5' position of codons. For example, NNG codons are preferred over synonymous NNA codons in the 5' position of lysine codons (NNG AAA) [74]. Mononucleotide repeats in coding sequence that result in transcriptional or translational slippage and frameshifting are avoided [69]. The dinucleotide bias at codon–codon junctions (cP3–cA1) influences codon pair frequencies, where 3rd base of a P-site codon (cP3) influences the choice of the first base of the A-site codon (cA1). The most frequently avoided type of cP3–cA1 dinucleotide or codon pairs contain the patterns NNUANN, NNGGNN, NNGNNC, NNCGCN, UUCGNN, CUCCNN, GUCCNN and NNCNNA. Meanwhile, the most frequently preferred codon pairs contain the patterns NNGCNN, NNCANN or NNUNCN in Bacteria, Archaea and eukaryotes [69]. In dicots, XCG is always the least favored codon. The G ending codons for Thr, Pro, Ala and Ser are avoided in both monocots and dicots because they contain C in codon position 2. UpA dinucleotides are avoided in the three domains of life (Archaea, prokaryotes and eukaryotes), while CpG dinucleotides are rejected in higher eukaryotes. However, UpG and CpA dinucleotides were strongly preferred in higher eukaryotes and ApA and UpU dinucleotides overall preferred (due to occurrence of three or more identical bases in all three domains of life). In plant genomes, a general bias exists in the use of specific dinucleotides and trinucleotides in different genomic regions [75].
The nucleotide composition surrounding the start codon AUG is significantly biased. 'A/G’ and 'G’ nucleotides were respectively preferred preceding and just following the start codon AUG (A/G AUG G), while a 'C’ following AUG was less favored in the Kozak sequence of the translation start site [71]. AUG codon context is quite diverse among different species and consensus sequence of AUG context for high translational efficiency in plants does not conform to the Kozak sequence found in animal systems. Most frequent nucleotides around the initiation site was reported to be A(A/C)AAA + 1UGGC in eudicots and A(A/G)CCA + 1UGGC in monocots. There is preference for G in position + 4 (85%) and C at + 5 (77%) in plants [76]. Preference and restrictions also exist for stop codons. At the 3' side of translation termination codon, uridine is the nucleotide most frequently found. Context of stop codons has conserved sequence patterns and thus termination signals may contain more than four nucleotides. In seven eukaryotic classes, significant differences were revealed in stop-codon context at first position in the downstream region (+ 1) and last two positions in the upstream region (− 1 and − 2) [77]. The codons resembling stop codons (URR) are restricted in highly expressed genes to prevent premature termination of translation. GC content of genome and RF1/RF2 ratio have a strong impact on stop codon frequencies, where RF1 is release factor 1 for UAA and UAG and RF2 is the release factor 2 for UAA and UGA. RF1/RF2 abundance ratio is linked to the ratio between number of genes with UAG and UGA stop codons. In bacteria, the frequency of UAA and UGA stop codons strongly depends on the genomic GC-content, but UAG frequency is independent of the GC content. When GC content is high, UGA is the preferred stop codon. In highly expressed genes, UAA is more frequent or optimal stop codon, while UAG is suboptimal codon. The sequences UAAUG, UGAUG and AUGA (stop codons are underlined) are excluded to remove potential overlapping start- and stop codons [78]. On the other hand, in eukaryotes, UGA was the most frequent stop codon, UAA was intermediate and UAG the least frequent in terms of usage [71]. In plants, the preference is in the order UGA > UAA > UAG [77].
Intergenic codon bias
Codon bias occurs not only within individual genes (intragenic) but also between genes, that are either clustered in operons or scattered in a genome. A highly stable, non-random dinucleotide frequency pattern identified in bulk genomic DNA is called general design. The relative abundance of dinucleotides constitute a ‘genome signature’ [70, 79]. In mammals, bacteria and plants, several dinucleotide sequences such as CpG, TpA and GpA are under-represented and hence the first nucleotide after each codon drives synonymous codon choice. Codon choice also maintains the efficiency of global protein translation in the cell. Intergenic codon bias in genes is randomly distributed in the genome and facilitates their differential expression. Codon choice of some genes would affect the translation of others due to a “shared economy” of the entire translation apparatus. Hence, codon usage also acts in ‘trans’ [12]. Functionally related genes show similar codon bias patterns for their co-regulation under specific conditions and codon optimality helps to coordinate their expression patterns. However, degree of codon bias of related genes may significantly differ as in the genes within prokaryotic operons [1]. In yeast, proteins involved in metabolically important processes such as glycolysis are enriched with optimal codons, while the regulatory products are enriched with non-optimal codons so that, response to transient stimuli or stress can be curtailed, when the stimulus is withdrawn [80].
tRNA abundance and tRNA interactions
Codon bias in relation to tRNA can be categorized into (a) Frequency bias, where the frequency of used codons matches the cellular tRNA population, (b) Co-occurrence bias, where synonymous codons recognized by same tRNA, cluster together and (c) Pair bias, which is a bias for optimal interactions of tRNAs in the A and P sites of the ribosomes, during translation [1].
The most ‘preferred codons are those for which the respective tRNAs are abundant [10]. The single major codon in synonymous group is complementary to the most abundant iso-accepting tRNA, which is termed as frequency bias. In many prokaryotes and eukaryotes, tRNA abundance correlates with codon usage and amino acid composition. In multicellular organisms, this relation between codon frequency bias and tRNA abundance is measured using tRNA adaptation index (tAI), which is based on the copy number of tRNA genes and efficiency of codon-anticodon binding [1]. Use of codons that match the most abundant tRNA, reduces the time to find and bind the correct tRNA, decreasing the likelihood of binding a non-cognate tRNA. In plants, tRNA population functionally adapts to codon frequency. The codon usage pattern in zein protein in maize was found to fit well with the array of most abundant tRNA iso-acceptors in the endosperm [70]. However, positive correlation between codon usage and tRNA abundance did not occur in many instances. In multicellular organisms with larger genomes, higher tRNA gene redundancy decreased selection for specific codons [1]. Certain amino acid sequences that are required for protein function and conformation do not use most abundant tRNAs. In highly expressed genes of rice, correspondence between majority of preferred codons and tRNA gene copy number was observed in GC-poor class, but not with GC-rich group of genes. On the other hand, the synonymous codon usage pattern in Arabidopsis was completely influenced by tRNA gene copy number. This was due to a huge variation of GC content in homologous gene sets in rice, which was absent in Arabidopsis [81]. tRNA gene number is negatively correlated with amino acid size or complexity, to minimize the use of large or complex amino acids that cause protein misfolding. Frequent and exclusive use of abundant tRNAs for most genes produces a shortage of these tRNAs within a cell, which is a mechanism to accommodate other more important genes [50].
Wobble or non-Watson–Crick base-pairing (such as G-U at the third position of a codon, corresponding to the 5′ position of the anticodon of a tRNA) and modification of nucleotides in tRNAs, extend the range of recognized synonymous codons. In bacteria, tRNA modifications of uridine-34 to hydroxy-uridine and derivatives allow wobble pairing with A, G and U, while in eukaryotes and few bacteria, tRNA modification of adenine-34 to inosine-34, allows wobble pairing with A, C and U. Also, synonymous codons recognized by the same tRNA are clustered (codon co-occurrence bias) to ensure interaction of tRNAs in the A and P sites of the ribosome to enhance translation efficiency [1]. Certain codons would be less likely to occur next to each other when particular tRNAs interfere or are incompatible when brought close together on the ribosome [82]. This also explains how tRNA-tRNA interactions affect codon usage.
tRNA-ribosome interactions also could be reflected in codon usage. Confirmational flexibility of L-shaped structure of tRNA, along with confirmational changes and rearrangements of the ribosomes, respectively provide variety of tRNA binding states and ability of ribosome to bind to tRNA hybrid states [83]. After their exit from the E site, tRNAs remaining near translating ribosome, are recharged by the corresponding amino-acyl-tRNA synthetases and when the same or iso-accepting codon occurs, these tRNAs start translation in the ribosome [1]. Ribosomal dwelling time or ribosomal occupancy on a codon is determined by codon usage as well as amino acid context of codon. If ribosomes traverse optimal codons rapidly, then ribosomes will be scarce when the ribosomal-A site is positioned over optimal codons. Treatment of cells with translation elongation inhibitor cycloheximide showed that tRNA abundance and codon-level ribosome density were not correlated. However, in cycloheximide-free systems, tRNA abundance and ribosome occupancy was found to be inversely related, indicating that ribosomes spend less time at optimal codons [80]. Optimal codons allow rapid ribosome translocation, while stretches of non-optimal codons or inhibitory codon pairs and their synergestic action slows ribosome translocation, since ribosomes wait for a rare cognate tRNA, eventually creating ribosome crowding, that can inhibit translation initiation. Such inhibitory codon pairs had at least a codon which interacted with its cognate tRNA via wobble pairing and exhibited long ribosomal dwelling times at P and A or E and P ribosomal sites. Slowing or increased dwelling time of ribosomes result when non-optimal codon doublets occur or when a codon-tRNA cognate pair is rare, or when a codon must be decoded by wobble tRNAs. Also, a decrease in (aminoacyl) tRNA availability due to starvation or shortage of amino acids, increases ribosome stalling [80].
mRNA secondary structure
RNA level selection acts on synonymous sites in both prokaryotes and eukaryotes. Synonymous codon positions define mRNA secondary structure and stability, translation rate and folding as well as post-translational modifications of nascent polypeptides [49]. Single-stranded mRNA molecules form secondary structures through complementary self-interactions. Periodic nucleotide patterns created by the genetic code as well as synonymous codon usage and relative abundance of dinucleotides are involved in mRNA secondary structures. A gene is more stable when folded as RNA or DNA, and this secondary structure protects the functionally important sites from detrimental mutations and decreases the evolutionary variability [84]. Functional domains of the mRNA (CDS and 5′and 3′-UTRs) preferentially fold onto themselves, while the start codon and stop codon regions have relaxed secondary structures. Synonymous codons are selected to maintain a more stable and ordered mRNA secondary structure [49]. Higher mRNA structure improves interaction with RNA binding proteins (RBPs) that positively impacts translation or reduces accessibility to single strand-specific endonucleases improving functional mRNA half-life [85]. Synonymous sites can vary because of redundancy in genetic code, but messenger RNA secondary structure restricts this freedom [84]. Constraints acting on mRNA secondary structure were responsible for modulating codon usage variations in rice tissue-specific genes [81].
Synonymous codon usage bias favors AT-richness at third codon nucleotide positions. High sequence variability, particularly at third codon positions (or synonymous sites), inversely correlates with mRNA stability and affects mRNA secondary structures such as helices or loops. A high C content at third codon sites increases the number of potential G:C base-pairs, which are stronger than A:U interactions. G:C pairings make helices more bendable. Mutations at third codon position nucleotides in helices are selected against so that the helix-forming regions accumulate lesser mutations than loops. Synonymous changes that increase CpG extends mRNA half-life in vitro while ApU increase results in mRNA degradation. An increase in 3rd position Cs reflects mRNA secondary structure and stability. In both α-helices and β-sheets of protein secondary structures that are preferentially coded by mRNA stems, G is more abundant than C at first and second non-synonymous sites. Cytosine is preferred at third sites to maintain stable stems in these regions [84, 86]. Housekeeping genes have unusually low rates of protein evolution, their mRNAs have unusually high relative stability [86]. Transcript length controls secondary structure stability because mutations of mRNAs results in destabilization of longer mRNAs but not of shorter mRNAs [84].
Gender specificity, mating system and effective population size
Gender-specific selective pressures on codon usage could alter gene evolution and structure, influencing reproductive biology. Genes expressed in female had greater CUB than in male organs and gametes and exhibited greater usage of species-specific preferred codons. However, highly expressed genes have greater codon bias than lowly expressed genes, irrespective of gender specificity [87]. Mating system also affects codon usage, where efficacy of selection on non-synonymous mutations is reduced in a highly inbred species relative to outcrossed sibling species [88]. Codon bias is determined by mutation, genetic drift and natural selection on efficiency of translation. Selection on codon usage is strong in species with large effective population sizes. Codon bias declines with reduction in effective population size and long-term reduction leads to major shift in genome evolution. When effective population size is small, genetic drift becomes dominant over natural selection. Self-fertilisation reduces effective population size and reduction is more in organisms with haploid than for diploid selfing. In Physcomitrella patens with AT rich genome and showing haploid selfing, optimal codon usage and GC content are low [7, 89]. Closely related species do not usually exhibit major shifts in codon preferences, but changes in mutation rates over short time scales are quite common in large effective population sizes [7].
Codon usage in nuclear and organelle genes of plants
Transformation of crop plants with foreign genes is mainly achieved by targeting the desired genes at the nucleus. Hence the study of codon usage in nuclear genes of plants assumes importance. For functionally homologous genes, codon usage differs across species. The synonymous codon usage of nuclear genes of plants varies between monocots and dicots. In Graminae (monocot) genes, a gradient of GC content and codon usage existed along with the direction of transcription. The 5' ends of monocot genes were up to 25% more rich in GC content than their 3' ends, but not in dicot genes [90]. Codon usage variation in monocots is mainly determined by spatial arrangement of genomic G + C-content, or the isochore structure [81]. The silent third nucleotide position of codons is GC-rich in the monocot genomes (59–61% of G + C content), but AU-rich in the eudicot genomes (35–42% of G + C content) [45]. Monocot genes can be classified into those with narrow codon bias (high G3 + C3 values) and broader codon bias (lower G3 + C3). More biased codon usage is seen in highly expressed genes and more random usage in low-expression genes. The CpG intercodon dinucleotides are few or under- represented, frequently methylated and scattered in both monocots and dicots. However, TpG is over-represented. In Graminae, amino acid specific behavior is seen in codon usage, where T-ending codons were preferred for glycine and alanine [70]. Codon usage gradients were strongest for aminoacids with largest number of synonymous codons. Codon usage pattern in gymnosperm Ginko biloba tended towards A/U-ending codons, which showed an obvious gradient progressing from gymnosperms to dicots to monocots [58].
Plastids and mitochondria are thought to be prokaryotes in symbiotic association with a eukaryotic cell, during evolution. The codon usage of organellar genes is therefore more similar to that of prokaryotes. Plastid genome is small and encodes a limited set of genes fully expressed within the organelle [91]. During plant evolution, some plastid genes have moved to the nuclear genome, adjusted their base composition to nuclear genes, expressed in the nucleus and their products were transported to chloroplasts. The G + C content of such genes also increased when they integrated into the nuclear genome. The average GC content of entire genes, and at the three codon positions individually, was higher in nuclear than in chloroplast genes, in four angiosperm species (rice, maize, wheat and Arabidopsis), suggesting different genomic organization and mutation pressures in nuclear and chloroplast genes. Codon usage pattern of chloroplast genes differed from nuclear genes by their AU-richness and bias towards NNA and NNU codons, whereas G, C or U-ending codons were optimal in nuclear genomes [91, 92]. Chloroplast genes have low CUB and lower GC than AT content. Chloroplast genomes of Asteracea family had a narrow GC distribution without significant correlation between GC12 and GC3, and purines were used more frequently than pyrimidines [93]. Natural selection might have played a prominent role over mutation pressure in sculpturing the CUB of chloroplast genes [94].
Codon usage bias at a particular site is influenced by flanking codons, composition of 3ʹ flanking nucleotide and amino acid content. In highly expressed plastid genes with a high overall codon adaptation, codon adaptation is lower particularly within the 1st 25 codons, at the 5' end of genes [67]. Strong context-dependent codon bias was observed in chloroplasts of flowering plants [91]. Highly expressed genes show an overall bias towards the NNC codons, which is strongest upstream of a C, but weakest upstream of a G. When the 3' neighboring base is a G, the bias changes towards NNT [67]. The bias towards NNC codons is to avoid CpG sites in coding region, but CpG is not generally avoided in plastid non-encoding regions. Avoidance of CpG is not a general compositional feature but is specific to NNY groups. While CpG methylation influences codon bias in the nuclear genome, CpG methylation is rare in chloroplast genomes. CpG is not methylated in plastid genes due to lack of methylase activity. However, CpGs are heavily modified in amyloplasts and chromoplasts indicating that plastid SCUB is also affected by DNA methylation [67, 95]. Plastid genes also have atypical start codons such as GTG, TTG, CAC, TTG ACG, ATC, ATT and TAC and atypical stop codons such as CAA, TCA, CGA, and CAG. The preference for typical stop codons is in the order TAA > TAG > TGA. Internal stop codons rarely exist in the coding sequence of plastid genome due to RNA editing or processing mechanism than converts U to C, which eliminates internal stop codons [95].
The codon usage in plastid genes is dependent on the aminoacid content. Highly expressed genes have increased proportions of certain amino acids since codons for these (G + C rich or GNN codons, in particular) are more efficiently translated [67]. SCUB patterns in chloroplast genomes were distinct based on ploidy level and reflected the polyploid formation from their diploid progenitors. Total frequency of SCUB did not vary between polyploids and diploids, but the SCUB for plastid coding sequences were distinct for polypoids and diploids or their progenitor species [96]. Synonymous codon usage bias (SCUB) is correlated with both intron number and exon position in the plant nuclear genome but not in the plastid genome [95]. In the nuclear and organellar genomes, the frequency of NNA/T codons rises as the intron number increases. However, in chloroplasts, NNC/G codons are preferred in genes with more introns. SCUB in exonic sequence was unaffected by polyploidization in Gossypium while heterogenity of SCUB prevailed in Triticum sp [96].
Organisms across all domains of life, never contain full set of tRNAs with anticodons complementary to the 61 different codons and the tRNA numbers may vary from 28 to 47 or more. A single tRNA can translate multiple synonymous codons through wobble base-pairing (G-U at the third codon position) and by super wobbling due to tRNA nucleotide modifications as described above (in tRNA abundance and interactions) [1]. Organelles also do not encode full set of tRNA species required to read all codons. In A. thaliana, O. sativa and P. trichocarpa chloroplast genomes, 37, 38 and 39 tDNAs have been annotated, respectively which correspond to 30 tRNA isoacceptor species. Chloroplast genomes encode most of the tRNAs required for translation, while missing tRNAs are imported from the cytosol. In Balanophora with an extremely AT-rich genome and AT-rich plastid protein genes, plastid genome (plastome) had no tRNAs and plastids imported all tRNAs required for translation [97]. Many plastid genes have been lost or transferred to the nucleus during evolution. The number of organellar tDNA insertions in nuclear genome varies from one plant species to another. None of plastid tRNA genes were found to be functional after integration to nuclear genome in five angiosperms (A. thaliana, M. truncatula, P. trichocarpa, O. sativa, B. distachyon) and green alga (C. reinhardtii) though tRNA genes maybe functional after integration [32].
In plant mitochondrial genomes, 17–29 tDNAs have been identified, except in Chlamydomonas mitochondria, which had only three tDNAs. Import of tRNAs from nucleus, plastid and cytosol compensates for the deficiency of mitochondrial tDNAs [32]. In plant mitochondria, codon usage patterns were more conserved in GC content, no correlation prevailed between GC12 and GC3, and T/A ending codons were preferred, the preference being more in genes with a greater number of introns, though the bias was also seen among exons and T was more frequently used than A [98].
Analysis of codon usage
Codon usage can be quantified in different ways. Earlier studies on synonymous codon usage were based on a sample of 100 genes from a genome. The first catalogue of codon usage frequencies was tabulated from mRNA sequences of 50 (or more) codons in length by Grantham and colleagues in 1980 [63]. As large volumes of sequence data were produced, surveys of codon usage required automation in extracting protein-coding DNA sequences from the primary databases and in subsequent statistical analysis of thousands of genes. DNA sequence information can be obtained directly from GenBank, EMBL or NCBI database. Numerous measures or a variety of indices have been developed since the 1980s to describe, analyse and quantify codon usage bias or codon use preferences [99]. The methods of analysis of codon usage bias can be split into 2 main categories viz., those that compare the observed codon usage distribution of target coding sequence against the reference set of highly-expressed genes and those that compare distribution based on assumption of uniform usage of synonymous codons [99]. Some most common methods used for analysis of codon usage bias (CUB) are given below.
Correspondence analysis
Correspondence analysis (CA) or factorial correspondence analysis (FCA) is a graphical two-dimensional representation of multivariate count or proportion data. Data are expressed as a matrix or two-way contingency table in which rows correspond to genes and columns show codons. A sample of G number of genes can be arranged with G rows and 61 columns [100]. CA can be used to extract the trends in the data set or trends among the genes either using raw counts (containing synonymous codon usage information) or counts corrected for amino acid usage or relative synonymous codon usage values.
Frequency of optimal codons (fop)
It is the ratio between the frequency of optimal codons and the total number of synonymous codons, and is a species-specific measure [47].
Effective number of codons
The effective number of codons (ENC or Nc) index is a measure of the extent of codon preference in a gene and quantifies the extent of departure of a gene from uniform or equal usage of synonymous codons within each amino acid class. ENC is best overall estimator of absolute synonymous codon usage biases and can be easily calculated from codon usage data alone. To investigate codon usage patterns across genes, ENC is plotted against factors such as GC3 to constitute ENC plot. ENC expected = 2 + s + (29s2 + (1 − s)2), where s indicates the frequency of GC3s [101].
For each gene, value of ENC lies between 20 (extreme bias when only one codon is used for each amino acid) and 61 (when all codons are uniformly used). ENC values ≤ 35 are indicative of genes with significant codon bias [101]. A higher ENC value indicates weaker codon usage bias [4]. ENC is independent of gene length and amino acid composition, does not rely on organism-specific data and can be easily applied to study new organisms.
Neutrality plot
Neutrality plot is used to analyse the effects of natural selection and mutation pressure on codon usage. The GC12 values are plotted against GC3 values, and a regression line is plotted. When slope of the regression curve is 1, codon usage bias is due to mutation pressure, and when the slope is towards 0, natural selection is considered the main force shaping codon usage [102].
Parity rule 2 (PR2) bias plot analysis
Parity rule 2 (PR2) plot analysis is used to evaluate the effect of mutation pressure and natural selection at the third codon position of the four-codon amino acids. The PR2 plot distinguishes between AU bias [A3/ (A3 + U3)] and GC bias [G3 / (G3 + C3)]. The AU bias and GC bias of each gene are calculated and AU bias is plotted against GC bias, to show the relationship between the contents of purines (A and G) and pyrimidines (T and C) at the third codon position of genes. At the centre of the plot where A = T and G = C (PR2) and both coordinates are 0.5, there is no deviation between mutation and selection pressure of two DNA chains (where A + T + G + C = 1) and the effect of mutation pressure and natural selection are equal. The degree of deviation from PR2 estimates the chain bias affected by mutation, selection, or both. If the codon usage frequency of A + T is the same as that of G + C at the third position, then the codon usage preference is likely to be entirely caused by mutation [102].
Codon adaptation index
Codon adaptation index or CAI is a simple, effective measure that shows the degree of codon usage bias towards the major codons. CAI for a specific gene can be determined by comparing its codon usage frequency with the reference set of highly expressed genes from a species. CAI score for a gene is calculated from the frequency of use of all codons in that gene. CAI value ranges between 0 and 1.0, and a higher CAI value means a stronger codon usage bias and a higher expression level [4]. Programs like Emboss chips may be used to calculate CAI value for the genes. CAI can be used to compare codon usage in different genes and organisms, predicting the expression level of a gene and indicates approximately the success of heterologous gene expression. CAI and ENC are the most popular indices. CAI measures the degree of bias towards a specified set of adaptive codons as opposed to ENC which measures only the degree of deviation from uniform codon usage, regardless of which codons are over-represented. Higher codon bias is indicated by lower ENC and higher CAI [91].
Relative synonymous codon usage
Relative synonymous codon usage (RSCU) is used to analyse codon usage variation between genes. RSCU value for a codon is the observed frequency of the codon divided by the expected frequency of the same codon within a synonymous codon group in the entire coding sequence of the gene, under the assumption of equal usage of the synonymous codons for an amino acid or in the absence of codon usage bias. Expected number of occurrences of a codon is ratio of the number of times the encoded amino acid is present in the protein sequence to the number of synonymous codons for the amino acid encoded by codon. An RSCU value of 1 shows no codon bias and greater than 1 means that a codon is used more often than expected, while values less than 1 indicate its relative rarity [9]. RSCU values can be 2, 3, 4 and 6 when a single codon is used to encode aminoacids having 2,3,4 and 6 synonymous codons respectively [51].
ARSCU or Average ratio of RSCU is a new index based on RSCU, which measures the ratio of RSCUs with GC-ending codons to the AT-ending codons for all amino acids in a gene.
Since RSCU may be zero for some codons, any RSCU with a value of zero is arbitrarily assigned a value of 0.1. ARSCU values was used to discriminate genes with different expressions under drought conditions, Genes with ARSCU above 13, 9 to 13 and less than 9 were predicted to be genes with high, high or intermediate and low or intermediate expression respectively, in rice under drought conditions [51].
Relative codon bias strength (RCBS)
RCBS is a codon bias index (CBI) to estimate codon bias without using a reference set.
L is the length of the gene represented in codons, RCBxyz is the RCB of codon xyz in the gene, f (x,y,z) the normalised observed frequency of codon xyz and f1(x), f2(y) and f3(z) are the normalised observed frequencies of bases x, y and z at codon positions 1, 2 and 3 respectively. For a particular codon, the observed codon frequency and then ratio of the observed to the expected codon frequency is calculated, which is derived as the product of the individual base frequencies at each codon position. RCBS of gene sequence is computed as the geometric mean of codon bias over all sequence codons. RCBS correlated significantly with CAI but was superior to CAI in predicting protein concentration and abundance. However, RCBS has a strong correlation with gene length, yielding larger values for shorter gene sequences which prevents its application to sequences shorter than 1000 bp. RCBS can be corrected with genomic pseudo-counts, but this in turn limits its application to complete genomes [103].
Modified relative codon bias strength or MRCBS is a score used to elucidate pattern and level of expression of genes in Arabidopsis [53]. MRCBS and CAI (codon adaptation index) were also strongly correlated. MRCBS relies exclusively on sequence features for identifying the highly expressed genes. MRCBS is also calculated in similar way as RCBS first for each codon and then for the whole gene.
RCBSaa, max is the maximum value of RCBS of codon encoding the same amino acid aa in the same reference set, and N is the codon length of the gene or query sequence. The score of MRCBS ranges from 0 and 1. The threshold score of MRCBS for identifying highly expressed genes, varies from genome to genome.
Relative codon adaptation (RCA)
Relative codon adaptation or RCA is a reference-set-based index similar to CAI and based on genomic base composition [103]. RCA uses a reference set to compute observed and expected codon frequencies.
L is the length of the query sequence, in codons, RCAxyz is the RCA of codon xyz, f (x,y,z) the observed frequency of codon xyz and f1(x), f2(y) and f3(z) the observed frequencies of bases x, y and z at codon positions 1, 2 and 3 respectively. Like CAI and RCBS, RCA is also computed as the geometric mean of the RCAxyz for each codon xyz in the sequence. However, unlike RCBS, RCA doesn’t have intrinsic bias for gene length, can be used for sequences shorter than 1000 bp and genomic corrections need not be applied. RCA outperforms CAI as a predictor of gene expression when operating on the CAI reference set.
Codon deviation coefficient (CDC)
CDC characterizes CUB, ascertains its statistical significance and requires no prior knowledge of reference gene sets. CDC takes into account both GC and purine contents, not only in sequences but also at three codon positions. It adopts the cosine distance metric to quantify CUB and employs bootstrapping to assess its statistical significance. CDC values range from 0 (no bias) to 1 (maximum bias) [99].
Other CUB indices and methods
Selection forces may directly shape the genome-wide relative frequency of codons and may operate at sequence level on individual genes (sequence level selection or SLS). A distance measure D was devised, that determines amount of bias in a particular genome, differences in the genome-wide frequency of codons and apparent non-random distributions of codons across mRNAs. Magnitude of D varies within taxonomic classes and its calculation requires no gene reference sets, so that it can be applied to poorly characterized genomes [104]. Relative codon deoptimisation index (RCDI) is another index which compares the similarity in codon usage of a given coding sequence with that of a reference genome. It can be used to measure host adaptability of plant viruses. An RCDI value of 1 indicates that the codon usage patterns are similar, while RCDI values higher than 1 indicates lower adaptability [99]. Within and across genomes, codon usage bias can be measured using the synonymous codon usage order (SCUO) purely in a mathematical way, based on the entropy of the amino acid in a sequence and with values varying from 0 to 1; a larger SCUO denoting a higher codon usage bias [105]. Gravy value is another measure which indicates the effect of protein hydrophobicity on codon usage bias with values ranging from − 2 to 2. On the other hand, aroma value measures the effect of aromatic hydrocarbon proteins on codon usage bias [101].
Softwares for codon usage analysis
A large number of codon usage bias indices have been devised to estimate and understand codon usage preferences. In many cases, computation and analysis is complex and not straight forward. Many softwares have been devised for ease of computation such as INteractive Codon usage Analysis or INCA, JCat (Java Codon Adaptation Tool), COUSIN (COdon Usage Similarity INdex) [14], Automated Codon Usage Analysis (ACUA) software, CAIcal [9] etc. Most of the softwares compute the CAI, ENC and occasionally other indices. COUSIN (http://cousin.ird.fr), includes COUSIN, seven other indices, and provides additional features such as statistical analyses, clustering, and codon usage optimization for gene expression [14]. Few softwares are updated, few work in Linux or UNIX such as codonW and few are obsolete or not available such as CodonExplorer. A list of online tools available for codon analysis is given in [9].
Databases of codon usage
The Codon Usage Database is a useful source of pre-calculated codon usage data. Kazusa codon database (https://www.kazusa.or.jp/codon/) tabulated using information from GenBank and High-performance Integrated Virtual Environment-Codon Usage Tables (HIVE-CUTs, hive.biochemistry.gwu.edu/review/codon), use publicly available sequencing database such as GenBank and NCBI’s RefSeq for analysing codon bias in plants. HIVE-CUTs is a more comprehensive tool that analyses codon usage between individual organisms and across taxonomical clade. CUB can be viewed, compared as well as graphically represented through commonly used indices [6]. The Codon Bias DataBase or CBDB, Microbial Genome Codon Usage Database, Prokaryotes Codon Usage Database etc. were developed for bacteria or prokaryotes [106].
Applications of codon usage studies
Analysis of codon usage facilitates the understanding of evolution, phylogenetic relations, host–pathogen co-evolution relationships and environmental adaptation of living organisms, molecular evolution of individual genes, horizontal gene transfer events between species, detection of protein coding regions in uncharacterized genomic DNA and translation studies [6, 105]. Knowledge of codon usage patterns can be utilised to design degenerate oligonucleotides for PCR amplification of a gene [46]. Codon usage bias may play a role in temporal or cyclic control of gene expression, protein structure and function, co-translational protein folding, recombinant protein production and protein functional classification [45]. In addition, consideration of G + C content or codon usage is important for high levels of gene expression.
The major application of codon usage data is in optimizing or redesigning a gene for high or optimum expression in heterologous systems. When a foreign gene is transferred into a crop, the codons in the gene are modified, to suit to the host plant codon usage pattern. Codon re-engineering is done for optimizing expression of heterologous proteins in plants. Optimal codons rather than overall abundant codons should be used for the optimization [45]. Several algorithms and softwares were developed for codon optimization and synthetic gene design such as Codon optimizer, OPTIMISER, Eugene, COStar, DNA-Tailor or D-Tailor, Codon Optimisation OnLine or COOL, Computationally Optimised DNA Assembly or CODA, ATGme, CodonWizard etc. [107,108,109]. Synthetic gene design involves designing candidate sequences by selecting codons at random, using their probabilities from the codon usage table, passing the sequences through filters to ensure other design criteria, then eliminate unfavorable codon pairs, extreme GC content, repetitive sequences, unfavourable mRNA structures etc., and finally including or excluding restriction sites as required [110].
Codons adapted to efficient elongation for endogenous genes will not correspond to those for heterologous genes, because overexpression causes amino acid starvation and alterations in the abundances of charged tRNA [10].
Codon optimization has been used in plants for increasing expression of ‘Cry’ proteins for pest resistance or recombinant proteins for molecular pharming. The cry genes of Bacillus thuringiensis and synthetic cry genes are transferred to crops for insect or pest resistance. A synthetic cry2AX1 gene (NCBI accession GQ332539.1) made with plant-preferred codons and expressed in rice and cotton showed significant protection against different types of lepidopteran insects [111,112,113]. However, designing of two kinds of codon optimised cry genes, one for monocot and another for dicot plants are also in vogue to improve the chances for getting events with desirable level of transgene expression. Wheat cytochrome P450 genes (with high GC content and strong codon usage bias) different from dicots were codon optimized (through recoding of the 5' end of genes with low usage codons using a single PCR mega primer) and introduced into yeast and tobacco, for use in bioremediation [114].
Codon optimization resulted in significant increases in gene expression (75- to 80-fold) to negligible enhancement. Increase of codon-optimized protein synthesis is at the translational level rather than on transcript abundance. Ribosome pause at certain codons is eliminated by codon optimization in chloroplasts. To express larger genes (greater than 200 kD) not only codon optimization, but also knowledge of tRNAs encoded by genome, compatibility with regulatory sequences for optimal translation initiation and elongation are important [13].
Use of non-canonical amino acids (ncAAs) through codon reassignment has been applied in analysis of protein structure and interaction, introduction of post-translational modifications, production of constrained peptides, antibody–drug conjugates and novel enzymes [115]. The scope of applications of codon usage analysis is thus immense.
Conclusions
Codon usage bias is a widely prevalent biological phenomenon observed across all life forms with significant biological functions, implications and applications. Codon bias is only one of the multitude factors affecting gene expression and codon usage is itself influenced by several factors in the biological system. Hence, understanding codon usage bias is not as simple as altering the DNA sequence or codons, but highly complex due to the difficulty in identifying or measuring the relative impact of the various factors or components. Though having significant application in plant biotechnology in terms of heterologous expression of foreign genes, codon usage bias is not thoroughly investigated in most crop plants, though general principles are known in prokaryotic systems. The computation or estimation of codon bias is as well complicated and technically challenging or demanding, thereby limiting our understanding of codon usage bias. Detailed analysis of this intriguing phenomenon in plants requires integration of statistical, computational and bioinformatics tools, simplification of the computational methods, and thorough study or delineation of factors influencing codon usage. This can further aid in devising better tools for synthetic gene design, and regulation of gene expression in heterologous gene expression systems, a major application of one of the most intricate ‘code within the genetic code’, the codon usage bias. Codon usage bias (CUB) is an example that shows that neither the genome nor the genetic code were designed or had evolved at random, but with a purpose.
Data Availability
Not Applicable. All data are presented in the manuscript.
Code availability
Not Applicable.
References
Quax TE, Claassens NJ, Söll D, van der Oost J (2015) Codon bias as a means to fine-tune gene expression. Mol Cell 59:149–161. https://doi.org/10.1016/j.molcel.2015.05.035
Ma QP, Li C, Wang J, Wang Y, Ding ZT (2015) Analysis of synonymous codon usage in FAD7 genes from different plant species. Genet Mol Res 14:1414–1422. https://doi.org/10.4238/2015.February.13.20
Liu Y (2020) A code within the genetic code: codon usage regulates co-translational protein folding. Cell Commun Signal 18:145. https://doi.org/10.1186/s12964-020-00642-6
Salim HMW, Cavalcanti ARO (2008) Factors influencing codon usage bias in genomes. J Braz Chem Soc 19:2. https://doi.org/10.1590/S0103-50532008000200008
Supek F (2016) The code of silence: Widespread associations between synonymous codon biases and gene function. J Mol Evol 82:65–73. https://doi.org/10.1007/s00239-015-9714-8
Athey J, Alexaki A, Osipova E, Rostovtsev A, Santana-Quintero LV, Katneni U, Simonyan V, Kimchi-Sarfaty C (2017) A new and updated resource for codon usage tables. BMC Bioinformatics 18:391. https://doi.org/10.1186/s12859-017-1793-7
Ingvarsson PK (2008) Molecular evolution of synonymous codon usage in Populus. BMC Evol Biol 8:307. https://doi.org/10.1186/1471-2148-8-307
Liu Q (2012) Mutational bias and translational selection shaping the codon usage pattern of tissue-specific genes in rice. PLoS ONE 7:e48295. https://doi.org/10.1371/journal.pone.0048295
Mazumdar P, Binti Othman R, Mebus K, Ramakrishnan N, Ann Harikrishna J (2017) Codon usage and codon pair patterns in non-grass monocot genomes. Ann Bot 120:893–909. https://doi.org/10.1093/aob/mcx112
Plotkin J, Kudla G (2011) Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 12:32–42. https://doi.org/10.1038/nrg2899
Zhou Z, Dang Y, Zhou M, Li L, Yu C-h, Fu J, Chen S, Liu Y (2016) Codon usage is an important determinant of gene expression levels largely through its effects on transcription. Proc Natl Acad Sci USA 113:E6117–E6125. https://doi.org/10.1073/pnas.1606724113
Frumkin I, Lajoie MJ, Gregg CJ, Hornung G, Church GM, Pilpel Y (2018) Codon usage of highly expressed genes affects proteome-wide translation efficiency. Proc Natl Acad Sci USA 115:E4940–E4949. https://doi.org/10.1073/pnas.1719375115
Kwon K-C, Chan H-T, León IR, Williams-Carrier R, Barkan A, Daniell H (2016) Codon optimization to enhance expression yields insights into chloroplast translation. Plant Physiol 172:62–77
Bourret J, Alizon S, Bravo IG (2019) COUSIN (COdon usage similarity INdex): a normalized measure of codon usage preferences. Genome Biol Evol 11:3523–3528. https://doi.org/10.1093/gbe/evz262
Crick FHC, Barnett L, Brenner S, Watts-Tobin RJ (1961) General nature of the genetic code for proteins. Nature 192:1227–1232. https://doi.org/10.1038/1921227a0
Nirenberg M (2004) Historical review: deciphering the genetic code—a personal account. Trends Biochem Sci 29:46–54
Stegmann UE (2016) ‘Genetic Coding’ reconsidered: an analysis of actual usage. Br J Philos Sci 67:707–730. https://doi.org/10.1093/bjps/axv007
Ling J, Söll D (2012) The genetic code: yesterday, today, and tomorrow. Resonance 17:1136–1142
Chatterjee S, Yadav S (2019) The origin of prebiotic information system in the peptide/RNA World: a simulation model of the evolution of translation and the genetic code. Life 9:25. https://doi.org/10.3390/life9010025
Saier MH Jr (2019) Understanding the genetic code. J Bacteriol 201:e00091-e119. https://doi.org/10.1128/JB.00091-19
Hamashima K, Kanai A (2013) Alternative genetic code for amino acids and transfer RNA revisited. Biomol Concepts 4:309–318. https://doi.org/10.1515/bmc-2013-0002
Koonin EV, Novozhilov AS (2009) Origin and evolution of the genetic code: the universal enigma. IUBMB Life 61:99–111. https://doi.org/10.1002/iub.146
Žihala D, Eliáš M (2019) Evolution and unprecedented variants of the mitochondrial genetic code in a lineage of green algae. Genome Biol Evol 11:2992–3007. https://doi.org/10.1093/gbe/evz210
Záhonová K, Kostygov AY, Ševčíková T, Yurchenko V, Eliáš M (2016) An unprecedented non-canonical nuclear genetic code with all three termination codons reassigned as sense codons. Curr Biol 26:2364–2369. https://doi.org/10.1016/j.cub.2016.06.064
Osawa S, Jukes TH, Watanabe K, Muto A (1992) Recent evidence for evolution of the genetic code. Microbiol Rev 56:229–264
Hamashima K, Mori M, Andachi Y, Tomita M, Kohara Y, Kanai A (2015) Analysis of genetic code ambiguity arising from nematode-specific misacylated tRNAs. PLoS ONE 10:e0116981. https://doi.org/10.1371/journal.pone.0116981
Mukai T, Lajoie MJ, Englert M, Söll D (2017) Rewriting the genetic code. Annu Rev Microbiol 71:557–577. https://doi.org/10.1146/annurev-micro-090816-093247
Pánek T, Žihala D, Sokol M, Derelle R, Klimeš V, Hradilová M, Zadrobílková E, Susko E, Roger AJ, Čepička I, Eliáš M (2017) Nuclear genetic codes with a different meaning of the UAG and the UAA codon. BMC Biol 15:8. https://doi.org/10.1186/s12915-017-0353-y
Noutahi E, Calderon V, Blanchette M, El-Mabrouk N, Lang BF (2019) Rapid genetic code evolution in green algal mitochondrial genomes. Mol Biol Evol 36:766–783. https://doi.org/10.1093/molbev/msz016
Lobanov AV, Turanov AA, Hatfield DL, Gladyshev VN (2010) Dual functions of codons in the genetic code. Crit Rev Biochem Mol Biol 45:257–265. https://doi.org/10.3109/10409231003786094
Yuan J, O’Donoghue P, Ambrogelly A, Gundllapalli S, Sherrer RL, Palioura S, Simonović M, Söll D (2010) Distinct genetic code expansion strategies for selenocysteine and pyrrolysine are reflected in different aminoacyl-tRNA formation systems. FEBS Lett 584:342–349. https://doi.org/10.1016/j.febslet.2009.11.005
Michaud M, Cognat V, Duchêne AM, Maréchal-Drouard L (2011) A global picture of tRNA genes in plant genomes. Plant J 66:80–93. https://doi.org/10.1111/j.1365-313X.2011.04490.x
Rother M, Krzycki JA (2010) Selenocysteine, pyrrolysine, and the unique energy metabolism of methanogenic Archaea. Archaea 2010:453642. https://doi.org/10.1155/2010/453642
Piatkov KI, Vu TTM, Hwang C-S, Varshavsky A (2015) Formyl-methionine as a degradation signal at the N-termini of bacterial proteins. Microb Cell Graz Austria 2:376–393
Cao X, Slavoff XA (2020) Non-AUG start codons: expanding and regulating the small and alternative ORFeome. Exp Cell Res. https://doi.org/10.1016/j.yexcr.2020.111973
Quast B, Mrusek D, Hoffmeister C, Sonnabend A, Kubick S (2015) Cotranslational incorporation of non-standard amino acids using cell-free protein synthesis. FEBS Lett 589:1703–1712
Vranova V, Rejsek K, Skene KR, Formanek P (2011) Non-protein amino acids: plant, soil and ecosystem interactions. Plant Soil 342:31–48. https://doi.org/10.1007/s11104-010-0673-y
Jin X, Park OJ, Hong SH (2019) Incorporation of non-standard amino acids into proteins: challenges, recent achievements, and emerging applications. Appl Microbiol Biotechnol 103:2947–2958. https://doi.org/10.1007/s00253-019-09690-6
Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV (2016) Ribosomal frameshifting and transcriptional slippage: from genetic steganography and cryptography to adventitious use. Nucleic Acids Res 44:7007–7078. https://doi.org/10.1093/nar/gkw530
Napthine S, Ling R, Finch LK, Jones JD, Bell S, Brierley I, Firth AE (2017) Protein-directed ribosomal frameshifting temporally regulates gene expression. Nat Commun 8:15582. https://doi.org/10.1038/ncomms15582
Depeiges A, Degroote F, Espagnol MC (2006) Translation initiation by non-AUG codons in Arabidopsis thaliana transgenic plants. Plant Cell Rep 25:55–61. https://doi.org/10.1007/s00299-005-0034-0
Pinheiro VB, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, Chaput JC, Wengel J, Peak-Chew SY, McLaughlin SH, Herdewijn P, Holliger P (2012) Synthetic genetic polymers capable of heredity and evolution. Science 336:341–344. https://doi.org/10.1126/science.1217622
Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Correa IR, Romesberg FE (2014) A semi-synthetic organism with an expanded genetic alphabet. Nature 509:385–388. https://doi.org/10.1038/nature13314
Deb B, Uddin A, Chakraborty S (2020) Codon usage pattern and its influencing factors in different genomes of hepadnaviruses. Arch Virol 165:557–570. https://doi.org/10.1007/s00705-020-04533-6
Wang L, Roossinck MJ (2006) Comparative analysis of expressed sequences reveals a conserved pattern of optimal codon usage in plants. Plant Mol Biol 61:699–710
Bailey-Serres J, Fennoy SI (1993) Synonymous codon usage in Zea mays L. nuclear genes is varied by levels of C and G-ending codons. Nucleic Acids Res 21:5294–5300
Xu C, Dong J, Tong C, Gong X, Wen Q, Zhuge Q (2013) Analysis of synonymous codon usage patterns in seven different Citrus species. Evol Bioinform Online 9:215–228. https://doi.org/10.4137/EBO.S11930
Camiolo S, Melito S, Porceddu A (2015) New insights into the interplay between codon bias determinants in plants. DNA Res 22:461–470. https://doi.org/10.1093/dnares/dsv027
Shabalina SA, Spiridonov NA, Kashina A (2013) Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res 41:2073–2094. https://doi.org/10.1093/nar/gks1205
Wang H-C, Hickey DA (2007) Rapid divergence of codon usage patterns within the rice genome. BMC Evol Biol. https://doi.org/10.1186/1471-2148-7-S1-S6
Chamani Mohasses F, Solouki M, Ghareyazie B, Fahmideh L, Mohsenpour M (2020) Correlation between gene expression levels under drought stress and synonymous codon usage in rice plant by in-silico study. PLoS ONE 15:e0237334. https://doi.org/10.1371/journal.pone.0237334
Barozai MY, Kakar A, Din M (2012) The relationship between codon usage bias and salt resistant genes in Arabidopsis thaliana and Oryza sativa. Pure Appl Bio 1:48–51. https://doi.org/10.19045/bspab.2012.12005
Sahoo S, Das SS, Rakshit R (2019) Codon usage pattern and predicted gene expression in Arabidopsis thaliana. Gene: X 2:100012. https://doi.org/10.1016/j.gene.2019.100012
Murray EE, Lotzer J, Eberle M (1989) Codon usage in plant genes. Nucleic Acids Res 17:477–498. https://doi.org/10.1093/nar/17.2.477
Ahmad T, Sablok G, Tatarinova TV, Xu Q, Deng XX, Guo WW (2013) Evaluation of codon biology in citrus and Poncirus trifoliata based on genomic features and frame corrected expressed sequence tags. DNA Res 20:135–150. https://doi.org/10.1093/dnares/dss039
Morton BR, Wright SI (2007) Selective constraints on codon usage of nuclear genes from Arabidopsis thaliana. Mol Biol Evol 24:122–129
Hui S, Jing L, Tao C, Zhi-biao N (2018) Synonymous codon usage pattern in model legume Medicago truncatula. J Integr Agric 17:2074–2081. https://doi.org/10.1016/S2095-3119(18)61961-6
He B, Dong H, Jiang C, Cao F, Tao S, Li-an Xu (2016) Analysis of codon usage patterns in Ginkgo biloba reveals codon usage tendency from A/U-ending to G/C-ending. Sci Rep 6:35927. https://doi.org/10.1038/srep35927
Chiapello H, Lisacek F, Caboche M, Henaut A (1998) Codon usage and gene function are related in sequences of Arabidopsis thaliana. Gene 209:GC1–GC38
Gonzalez A, Corsini G, Lobos S, Seelenfreund D, Tello M (2020) Metabolic specialization and codon preference of lignocellulolytic genes in the white rot basidiomycete Ceriporiopsis subvermispora. Genes 11:1227. https://doi.org/10.3390/genes11101227
Song H, Gao H, Liu J, Tian P, Nan Z (2017) Comprehensive analysis of correlations among codon usage bias, gene expression, and substitution rate in Arachis duranensis and Arachis ipaënsis orthologs. Sci Rep 7:14853. https://doi.org/10.1038/s41598-017-13981-1
Powell JR, Moriyama EN (1997) Evolution of codon usage bias in Drosophila. Proc Natl Acad Sci USA 94:7784–7790
Komar AA (2016) The Yin and Yang of codon usage. Hum Mol Genet 25(R2):R77–R85. https://doi.org/10.1093/hmg/ddw207
Clarke TF, Clark PL (2010) Increased incidence of rare codon clusters at 5’ and 3’ gene termini: implications for function. BMC Genomics 11:118. https://doi.org/10.1186/1471-2164-11-118
Bentele K, Saffert P, Rauscher R, Ignatova Z, Blüthgen N (2013) Efficient translation initiation dictates codon usage at gene start. Mol Syst Biol 9:675. https://doi.org/10.1038/msb.2013.32
Clarke TF 4th, Clark PL (2008) Rare codons cluster. PLoS ONE 3:e3412. https://doi.org/10.1371/journal.pone.0003412
Morton BR, So BG (2000) Codon usage in plastid genes is correlated with context, position within the gene, and amino acid content. J Mol Evol 50:184–193. https://doi.org/10.1007/s002399910020
Yurovsky A, Amin MR, Gardin J, Chen Y, Skiena S, Futcher B (2018) Prokaryotic coding regions have little if any specific depletion of Shine-Dalgarno motifs. PLoS ONE 13:e0202768. https://doi.org/10.1371/journal.pone.0202768
Tats A, Tenson T, Remm M (2008) Preferred and avoided codon pairs in three domains of life. BMC Genomics 9:463. https://doi.org/10.1186/1471-2164-9-463
De Amicis A, Marchetti S (2000) Intercodon dinucleotides affect codon choice in plant genes. Nuclei Acids Res 230:1025–1054
Feng C, Xu C-j, Wang Y, LiuW-l Yin X-r, Li X, Chen M, Chen K-s (2013) Codon usage patterns in Chinese bayberry (Myrica rubra) based on RNA-Seq data. BMC Genomics 14:732. https://doi.org/10.1186/1471-2164-14-732
Chu D (2021) Wei L Context-dependent and -independent selection on synonymous mutations revealed by 1,135 genomes of Arabidopsis thaliana. BMC Ecol Evo 21:68. https://doi.org/10.1186/s12862-021-01792-y
Moura G, Pinheiro M, Silva R, Miranda I, Afreixo V, Dias G, Freitas A, Oliveira JL, Santos MA (2005) Comparative context analysis of codon pairs on an ORFeome scale. Genome Biol 6:R28. https://doi.org/10.1186/gb-2005-6-3-r28
Morton BR (1998) Selection on the codon bias of chloroplast and cyanelle genes in different plant and algal lineages. J Mol Evol 46:449–459. https://doi.org/10.1007/PL00006325
Porceddu A, Camiolo S (2011) Spatial analyses of mono, di and trinucleotide trends in plant genes. PLoS ONE 6:e22855. https://doi.org/10.1371/journal.pone.0022855
Nakagawa S, Niimura Y, Gojobori T, Tanaka H, Miura K (2008) Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res 36:861–871
Sun J, Chen M, Xu J, Luo J (2005) Relationships among stop codon usage bias, its context, isochores, and gene expression level in various eukaryotes. J Mol Evol 61:437–444. https://doi.org/10.1007/s00239-004-0277-3
Belinky F, Babenko VN, Rogozin IB, Koonin EV (2018) Purifying and positive selection in the evolution of stop codons. Sci Rep 8:9260. https://doi.org/10.1038/s41598-018-27570-3
Karlin S, Burge C (1995) Dinucleotide relative abundance extremes: a genomic signature. Trends Genet 11:283–290. https://doi.org/10.1016/s0168-9525(00)89076-9
Hanson G, Coller J (2018) Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol 19:20–30. https://doi.org/10.1038/nrm.2017.91
Mukhopadhyay P, Basak S, Ghosh TC (2008) Differential selective constraints shaping codon usage pattern of housekeeping and tissue-specific homologous genes of rice and Arabidopsis. DNA Res 15:347–356. https://doi.org/10.1093/dnares/dsn023
Ernst JF (1988) Codon usage and gene expression. TIBTECH 6:196–199
Graifer D, Karpova G (2015) Interaction of tRNA with eukaryotic ribosome. Int J Mol Sci 16:7173–7194. https://doi.org/10.3390/ijms16047173
Krishnan NM, Seligmann H, Rao BJ (2008) Relationship between mRNA secondary structure and sequence variability in chloroplast genes: possible life history implications. BMC Genomics 9:48. https://doi.org/10.1186/1471-2164-9-48
Mauger DM, Cabral BJ, Presnyak V, Su SV, Reid DW, Goodman B, Link K, Khatwani N, Reynders J, Moore MJ, McFadyen IJ (2019) mRNA structure regulates protein expression through changes in functional half-life. Proc Natl Acad Sci USA 116:24075–24083. https://doi.org/10.1073/pnas.1908052116
Chamary J, Hurst LD (2005) Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol 6:R75. https://doi.org/10.1186/gb-2005-6-9-r75
Whittle C-A, Malik MR, Krochko JE (2007) Gender-specific selection on codon usage in plant genomes. BMC Genomics 8:169. https://doi.org/10.1186/1471-2164-8-169
Qiu S, Zeng K, Slotte T, Wright S, Charlesworth D (2011) Reduced efficacy of natural selection on codon usage bias in selfing Arabidopsis and Capsella species. Genome Biol Evol 3:868–880. https://doi.org/10.1093/gbe/evr085
Szövényi P, Ullrich KK, Rensing SA, Lang D, van Gessel N, Stenøien HK, Conti E, Reski R (2017) Selfing in haploid plants and efficacy of selection: codon usage bias in the model moss Physcomitrella patens. Genome Biol Evol 9:1528–1546. https://doi.org/10.1093/gbe/evx098
Wong GK, Wang J, Tao L, Tan J, Zhang J, Passey DA, Yu J (2002) Compositional gradients in Gramineae genes. Genome Res 12:851–856. https://doi.org/10.1101/gr.189102
Suzuki H, Morton BR (2016) Codon adaptation of plastid genes. PLoS ONE 11:e0154306. https://doi.org/10.1371/journal.pone.0154306
Wang Z, Xu B, Li B, Zhou Q, Wang G, Jiang X, Wang C, Xu Z (2020) Comparative analysis of codon usage patterns in chloroplast genomes of six Euphorbiaceae species. PeerJ 8:e8251. https://doi.org/10.7717/peerj.8251
Nie X, Deng P, Feng K, Liu P, Du X, You FM, Weining S (2014) Comparative analysis of codon usage patterns in chloroplast genomes of the Asteraceae family. Plant Mol Biol Rep 32:828–840. https://doi.org/10.1007/s11105-013-0691-z
Bhattacharyya D, Uddin A, Das S, Chakraborty S (2019) Mutation pressure and natural selection on codon usage in chloroplast genes of two species in Pisum L. (Fabaceae: Faboideae). Mitochondrial DNA A DNA Mapp Seq Anal 30:664–673. https://doi.org/10.1080/24701394.2019.1616701.a
Qi Y, Xu W, Xing T, Zhao M, Li N, Yan L, Xia G, Wang M (2015) Synonymous codon usage bias in the plastid genome is unrelated to gene structure and shows evolutionary heterogeneity. Evol Bioinform Online 11:65–77. https://doi.org/10.4137/EBO.S22566
Tian G, Li G, Liu Y, Liu Q, Wang Y, Xia G, Wang M (2020) Polyploidization is accompanied by synonymous codon usage bias in the chloroplast genomes of both cotton and wheat. PLoS ONE 15(11):e0242624. https://doi.org/10.1371/journal.pone.0242624
Su HJ, Barkman TJ, Hao W, Jones SS, Naumann J, Skippington E, Wafula EK, Hu JM, Palmer JD, dePamphilis CW (2019) Novel genetic code and record-setting AT-richness in the highly reduced plastid genome of the holoparasitic plant Balanophora. Proc Natl Acad Sci USA 116:934–943. https://doi.org/10.1073/pnas.1816822116
Zhou M, Li X (2009) Analysis of synonymous codon usage patterns in different plant mitochondrial genomes. Mol Biol Rep 36:2039–2046. https://doi.org/10.1007/s11033-008-9414-1
Zhang Z, Li J, Cui P, Ding F, Li A, Townsend JP, Yu J (2012) Codon Deviation Coefficient: a novel measure for estimating codon usage bias and its statistical significance. BMC Bioinformatics 13:43. https://doi.org/10.1186/1471-2105-13-43
Perrière G, Thioulouse J (2002) Use and misuse of correspondence analysis in codon usage studies. Nucleic Acids Res 30:4548–4555. https://doi.org/10.1093/nar/gkf565
He Z, Gan H, Liang X (2019) Analysis of synonymous codon usage bias in Potato Virus M and its adaption to hosts. Viruses 11:752. https://doi.org/10.3390/v11080752
Yu X, Liu J, Li H, Liu B, Zhao B, Ning Z (2021) Comprehensive analysis of synonymous codon usage patterns and influencing factors of porcine epidemic diarrhea virus. Arch Virol 166:157–165. https://doi.org/10.1007/s00705-020-04857-3
Fox JM, Erill I (2010) Relative codon adaptation: a generic codon bias index for prediction of gene expression. DNA Res 17:185–196. https://doi.org/10.1093/dnares/dsq012
Deng Y, de Lima HF, Kalfon J, Chu D, von der Haar T (2020) Hidden patterns of codon usage bias across kingdoms. J R Soc Interface 17:20190819. https://doi.org/10.1098/rsif.2019.0819
Angellotti MC, Bhuiyan SB, Chen G, Wan XF (2007) CodonO: codon usage bias analysis within and across genomes. Nucleic Acids Res 35:W132–W136. https://doi.org/10.1093/nar/gkm392
Hilterbrand A, Saelens J, Putonti C (2012) CBDB: the codon bias database. BMC Bioinformatics 13:62. https://doi.org/10.1186/1471-210513-62
Gould N, Hendy O, Papamichail D (2014) Computational tools and algorithms for designing customized synthetic genes. Front Bioeng Biotechnol 2:41. https://doi.org/10.3389/fbioe.2014.00041
Webster GR, Teh AY, Ma JK (2017) Synthetic gene design-the rationale for codon optimization and implications for molecular pharming in plants. Biotechnol Bioeng 114:492–502. https://doi.org/10.1002/bit.26183
Sen A, Kargar K, Akgü E, Mustafa C, (2020) Pınar Codon optimization: a mathematical programing approach. Bioinformatics 36:4012–4020. https://doi.org/10.1093/bioinformatics/btaa248
Gustafsson C, Govindarajan S, Minshull J (2004) Codon bias and heterologous protein expression. Trends Biotechnol 22:346–353
Jadhav MS, Sakthi AR, Balakrishnan N, Sudhakar D, Udayasuriyan V (2020) Study of expression of indigenous Bt cry2AX1 gene in T3 progeny of cotton and its efficacy against Helicoverpa armigera (Hubner). Braz Arch Biol Technol 63:e20180428. https://doi.org/10.1590/1678-4324-2020180428
Chakraborty M, Sairam Reddy P, Mustafa G, Rajesh G, Laxmi Narasu VM, Udayasuriyan V, Rana D (2016) Transgenic rice expressing the cry2AX1 gene confers resistance to multiple lepidopteran pests. Transgenic Res 25:665–678
Manikandan R, Balakrishnan N, Sudhakar D, Udayasuriyan V (2016) Transgenic rice plants expressing synthetic cry2AX1 gene exhibits resistance to rice leaffolder (Cnaphalocrosis medinalis). 3 Biotech 6:10. https://doi.org/10.1007/s13205-015-0315-4
Batard Y, Hehn A, Nedelkina S, Schalk M, Pallett K, Schaller H, Werck-Reichhart D (2000) Increasing expression of P450 and P450-reductase proteins from monocots in heterologous systems. Arch Biochem Biophys 379:161–169
Cui Z, Johnston WA, Kirill A (2020) Cell-free approach for non-canonical amino acids incorporation into polypeptides. Front Bioeng Biotechnol 8:1031. https://doi.org/10.3389/fbioe.2020.01031
Acknowledgements
Sujatha Thankeswaran Parvathy thanks Indian Council of Agricultural Research, New Delhi and Centre for Plant Molecular Biology, Tamil Nadu Agricultural University, Coimbatore for the support. The topic was part of the postgraduation (MSc) course work of Sujatha Thankeswaran Parvathy at TNAU, Coimbatore. We immensely thank the esteemed reviewers and editorial board of the journal for the invaluable, critical comments and suggestions which helped us to improve the manuscript.
Funding
No funding was received for the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
STP has performed the literature search, compilation, drafting, preparation or writing and revision of the manuscript. VU has provided valuable inputs in conceptualizing, correcting and revision of the manuscript. VPB has contributed to fine-tuning of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not Applicable.
Consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Parvathy, S.T., Udayasuriyan, V. & Bhadana, V. Codon usage bias. Mol Biol Rep 49, 539–565 (2022). https://doi.org/10.1007/s11033-021-06749-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06749-4