Abstract
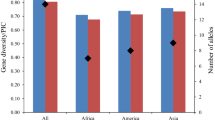
Wild soybeans, Glycine soja, are a source of genetic variation to improve soybeans. To improve the efficiency evaluation of conserved germplasm a core or mini-core collection approach that maximizes allelic diversity in a proportion of the whole collection has frequently been advocated. The genetic diversity of a wild soybean collection (1,305 accessions) plus Japanese cultivated soybeans (53 accessions) were analyzed at 20 SSR marker loci. Higher levels of allelic diversity were found in wild soybeans (28 alleles per locus) than Japanese cultivated soybean (five alleles per locus). The genetic distance between wild soybeans from different regions reflected their proximity. Accessions from Russia consisted of a diverse array of alleles resulting in accessions being spread further apart in a PCA plot than accessions from other regions. Accessions of wild soybean from Korea included many rare alleles and thus had a high representation in the core collection. The two core collections developed here, traditional and mini, consisted of 192 accessions with 97% of the allelic diversity (14% of the whole collection) and 53 accessions with 62.4% of the allelic diversity (5% of the whole collection), respectively.




Similar content being viewed by others
References
Abe J (2000) The genetic structure of natural populations of wild soybean revealed by isozymes and RFLPs of mitochondrial DNAs: possible influence of seed dispersal, cross-pollination and demography. In: K. Oono (ed) 7th MAFF International Workshop Genetic Resources, pp 143–158. AFFRC and NIAR, Tsukuba, Japan
Abe J, Hasegawa A, Fukushi H, Mikami T, Ohara M, Shimamoto Y (1999) Introgression between wild and cultivated soybeans of Japan revealed by RFLP analysis of chloroplast DNAs. Econ Bot 53:285–291
Abe J, Xu DH, Suzuki Y, Kanazawa A, Shimamoto Y (2003) Soybean germplasm pools in Asia revealed by nuclear SSRs. Theor Appl Genet 106:445–453
Brown AHD (1989) Core collections: a practical approach to genetic resources management. Genome 31:818–824
Brown AHD, Grace JP, Speer SS (1987) Designation of a ‘core’ collection of perennial Glycine. Soybean Genet Newsl 14:59–70
Excoffier L, Smouse P, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Gizlice Z, Carter TE, Burton JW (1994) Genetic base for North American public soybean cultivars released between 1947 and 1988. Crop Sci 34:1143–1151
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html. Updated from Goudet (1995). Accessed Mar 2009
Hodgkin T, Brown AHD, van Hintum ThJL, Morales EAV (eds) (1995) Core collections of plant genetic resources. Wiley, Chichester, p 269
Hyten DL, Song Q, Zhu Y, Choi YI, Nelson RL, Costa JM, Specht JE, Shoemaker RL, Cregan PB (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA 103:16666–16671. doi:10.1073/pnas.0604379103
Jin Y, He T, Lu BR (2003) Fine scale genetic structure in a wild soybean (Glycine soja) population and the implications for conservation. New Phytol 159:513–519. doi:10.1046/j.1469-8137.2003.00824.x
Kuroda Y, Kaga A, Tomooka N, Vaughan DA (2006) Population genetic structure of Japanese wild soybean (Glycine soja) based on microsatellite variation. Mol Ecol 15:959–974. doi:10.1111/j.1365-294X.2006.02854.x
Kuroda Y, Kaga A, Tomooka N, Vaughan DA (2008) Geneflow and genetic structure of wild soybean (Glycine soja) in Japan. Crop Sci 48:1071–1079. doi:10.2135/cropsci2007.09.0496
Lee J-D, Yu J-K, Hwang Y-H, Blake S, So Y-S, Lee G-J, Nguyen HT, Shannon JG (2008) Genetic diversity of wild soybean (Glycine soja Sieb. & Zucc.) accessions from South Korea and other countries. Crop Sci 48:606–616. doi:10.2135/cropsci2007.05.0257
Liu K, Muse S (2005) PowerMarker: new genetic data analysis software. Version 3.23, 2005. Available at http://powermarker.net. Accessed Mar 2009
Meilleur BA, Hodgkin T (2004) In situ conservation of crop wild relatives. Biodivers Conserv 13:663–684. doi:10.1023/B:BIOC.0000011719.03230.17
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Tajima N, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19:153–170. doi:10.1007/BF02300753
Powell W, Morgante M, Andre C, McNicol JW, Machray GC, Doyle JJ, Tingey SV, Rafalski JA (1995) Hypervariable microsatellites provide a general source of polymorphic DNA markers for the chloroplast genome. Curr Biol 5:1023–1029. doi:10.1016/S0960-9822(95)00206-5
Powell W, Morgante M, Doyle JJ, McNicol JW, Tingey SV, Rafalski JA (1996) Genepool variation in genus Glycine subgenus Soja revealed by polymorphic nuclear and chloroplast microsatellites. Genetics 144:793–803
Priolli RHG, Mendes-Junior CT, Arantes NE, Contel EPB (2002) Characterization of Brazilian soybean cultivars using microsatellite markers. Genet Mol Biol 25:185–193
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Qiu L, Guan R, Li Y, Wang L, Guan Y, Zhe Y, et al. (2004) The basis of sustainable soybean production: establishment and application of Chinese soybean core collections revealed by both agronomic characters and SSR markers. Fourth International Crop Science Congress (online)
Rohlf FJ (2000) NTSYS PC, version 2.02j. Exeter Software, Setauket
Sangiri C, Kaga A, Tomooka N, Vaughan DA, Srinives P (2007) Genetic diversity of the mungbean (Vigna radiata, Leguminosae) genepool based on microsatellite analysis. Aust J Bot 55:837–847. doi:10.1071/BT07105
Schneider S, Roessli D, Excoffier L (2000) Arlequin: a software for population genetic data. Genetics and Biometry Laboratory, University of Geneva, Switzerland
Shimamoto Y, Fukushi H, Abe J, Kanazawa A, Gai JY, Gao Z, Xu DH (1998) RFLPs of chloroplast and mitochondrial DNA in wild soybean, Glycine soja, growing in China. Genet Resour Crop Evol 45:433–439. doi:10.1023/A:1008693603526
Singh RJ, Nelson RL, Chung GH (2007) Soybean (Glycine max (L.) Merr.). In: Singh RJ (ed) Genetic resources, chromosome engineering, and crop improvement. Oilseed crops, vol 4. CRC Press, Boca Raton, pp 13–50
Takezaki N, Nei M (1996) Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389–399
Tozuka A, Fukushi H, Hirata T, Ohara M, Kanazawa A, Mikami T, Abe J, Shimamoto Y (1998) Composite and clinal distribution of Glycine soja in Japan revealed by RFLP analysis of mitochondrial DNA. Theor Appl Genet 96:170–176. doi:10.1007/s001220050724
Upadhyaya HD, Ortiz R (2001) A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement. Theor Appl Genet 102:1292–1298. doi:10.1007/s00122-001-0556-y
Xu DH, Abe J, Gai JY, Shimamoto Y (2002) Diversity of chloroplast DNA SSRs in wild and cultivated soybeans: evidence for multiple origins of cultivated cultivated soybeans. Theor Appl Genet 105:645–653. doi:10.1007/s00122-002-0972-7
Yaklich RW, Helm RM, Cockrell G, Herman EM (1999) Analysis of the distribution of the major soybean seed allergens in a core collection of Glycine max accessions. Crop Sci 39:1444–1447
Zhao L, Dong Y, Liu B, Hao S, Wang K, Li X (2005) Establishment of a core collection for the Chinese annual wild soybean (Glycine soja). Chin Sci Bull 50:989–996. doi:10.1360/982004-657
Acknowledgements
The authors thank the USDA for furnishing germplasm of wild soybeans from China, the Republic of Korea and Russia. The authors also thank the following research centers in Japan for supply of some of the cultivated soybean germplasm used in this experiment: Tokachi Agricultural Experiment Station, National Agricultural Research Center for Tohoku Region, Chushin Agricultural Experiment Station, National Agricultural Research Center for Kyushu Okinawa Region and Saga Agricultural Experiment Station.
Author information
Authors and Affiliations
Corresponding author
Additional information
All authors contributed equally to this research.
Rights and permissions
About this article
Cite this article
Kuroda, Y., Tomooka, N., Kaga, A. et al. Genetic diversity of wild soybean (Glycine soja Sieb. et Zucc.) and Japanese cultivated soybeans [G. max (L.) Merr.] based on microsatellite (SSR) analysis and the selection of a core collection. Genet Resour Crop Evol 56, 1045–1055 (2009). https://doi.org/10.1007/s10722-009-9425-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-009-9425-3