Abstract
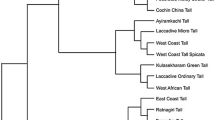
Simple sequence repeat (SSR) markers have been widely used to evaluate genetic resources and assist crop breeding in plant systems: they are codominant, easy to score and have abundant polymorphisms. In this study, we used coconut transcriptome sequences to determine the distribution characteristics of gene-based microsatellite loci in Cocos nucifera L. and to develop a series of polymorphic SSR markers. A total of 6,608 SSRs were identified across 57,304 unigenes from coconut transcripts: approximately one expressed sequence tag-derived SSR locus per 10 unigenes. We developed 309 primer pairs and characterized 191 polymorphic gene-based SSR markers across a set of ten coconut accessions from different countries. A total of 615 alleles were identified from the 191 polymorphic SSR loci, with an average of 3.22 alleles per locus. These gene-based SSR markers displayed moderate levels of polymorphism, with observed heterozygosity ranging from 0.18 to 0.695 (mean = 0.385 ± 0.14). Subsequently, 80 polymorphic markers were selected to assay genotypes of 82 coconut accessions collected from different regions in Asia. Association analysis found a total of nine markers significantly associated with coconut height (P < 0.05) using general linear models in a selective unstructured population and mixed linear models in a mixed-structure population. Two SSR markers (from Unigene9331 and Unigene3350) were significantly associated with coconut height using both statistical methods.



Similar content being viewed by others
References
Batugal PV, Rao R, Oliver J (2005) Coconut genetic resources. International Plant Genetic Resources Institute–Regional Office for Asia, the Pacific and Oceania (IPGRI-APO), Serdang, Selangor DE, Malaysia
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99(2):125–132
Fan H, Xiao Y, Yang Y, Xia W, Mason AS, Xia Z, Qiao F, Zhao S, Tang H (2013) RNA-Seq analysis of Cocos nucifera: transcriptome sequencing and de novo assembly for subsequent functional genomics approaches. PLoS One 8(3):e59997
Feng SP, Li WG, Huang HS, Wang JY, Wu YT (2009) Development, characterization and cross-species/genera transferability of EST-SSR markers for rubber tree (Hevea brasiliensis). Mol Breed 23(1):85–97
Flint-Garcia SA, Thornsberry JM, Edward S, Buckler IV (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54:357–374
Gunn BF, Baudouin L, Olsen KM (2011) Independent origins of cultivated coconut (Cocos nucifera L.) in the Old World Tropics. PLoS One 6(6):e21143
Gupta PK, Rustgi S, Kulwal PL (2005) Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol Biol 57(4):461–485
Gwaze DP, Zhou Y, Reyes-Valdes MH, Al-Rababah MA, Williams CG (2003) Haplotypic QTL mapping in an outbred pedigree. Genet Res 81:43–50
Jia G, Huang X, Zhi H, Zhao Y, Zhao Q, Li W, Chai Y, Yang L, Liu K, Lu H et al (2013) A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat Genet 45:957–961
Kantety RV, Rota ML, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48:501–510
Kaur S, Cogan NO, Pembleton LW, Shinozuka M, Savin KW, Materne M, Forster JW (2011) Transcriptome sequencing of lentil based on second-generation technology permits large-scale unigene assembly and SSR marker discovery. BMC Genom 12:265
Kraakman ATW, Niks RE, Van den Berg PMMM, Stam P, Van Eeuwijk FA (2004) Linkage disequilibrium mapping of yield and yield stability in modern spring barley cultivars. Genetics 168(1):435–446
Lawson MJ, Zhang L (2008) Housekeeping and tissue-specific genes differ in simple sequence repeats in the 5′-UTR region. Gene 407:54–62
Li Y-C, Korol AB, Fahima T, Nevo E (2004) Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 21(6):991–1007
Li X, Yan W, Agrama H, Jia L, Jackson A, Moldenhauer K, Yeater K, McClung A, Wu D (2012) Unraveling the complex trait of harvest index with association mapping in rice (Oryza sativa L.). PLoS One 7(1):e29350
Low E-TL, Alias H, Boon S-H, Shariff EM, Tan C-YA, Ooi LCL, Cheah S-C, Raha A-R, Wan K-L, Singh R (2008) Oil palm (Elaeis guineensis Jacq.) tissue culture ESTs: identifying genes associated with callogenesis and embryogenesis. BMC Plant Biol 8(1):62
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30(2):194–200
Palmieri DA, Novelli VM, Bastianel M, Cristofani-Yaly M, Astúa-Monge G, Carlos EF, de Oliveira AC, Machado MA (2007) Frequency and distribution of microsatellites from ESTs of citrus. Genet Mol Biol 30(3):1009–1018
Patel J (1937) Coconut breeding. Proc Assoc Econ Biol 5:1–16
Poncet V, Rondeau M, Tranchant C, Cayrel A, Hamon S, de Kochko A, Hamon P (2006) SSR mining in coffee tree EST databases: potential use of EST–SSRs as markers for the Coffea genus. Mol Genet Genomics 276(5):436–449
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Qiu D, Morgan C, Shi J, Long Y, Liu J, Li R, Zhuang X, Wang Y, Tan X, Dietrich E, Weihmann T, Everett C, Vanstraelen S, Beckett P, Fraser F, Trick M, Barnes S, Wilmer J, Schmidt R, Li J, Li D, Meng J, Bancroft I (2006) A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor Appl Genet 114(1):67–80
Qiu L, Yang C, Tian B, Yang J-B, Liu A (2010) Exploiting EST databases for the development and characterization of EST-SSR markers in castor bean (Ricinus communis L.). BMC Plant Biol 10(1):278
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA 98(20):11479–11484
Rivera R, Edwards KJ, Barker JHA, Arnold GM, Ayad G, Hodgkin T, Karp A (1999) Isolation and characterization of polymorphic microsatellites in Cocos nucifera L. Genome 42(4):668–675
Rostoks N, Ramsay L, MacKenzie K, Cardle L, Bhat PR, Roose ML, Svensson JT, Stein NT, Varshney RK, Marshall DF, Graner A, Close TJ, Waugh R (2006) Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. Proc Natl Acad Sci USA 103(49):18656–18661
Rozen S, Skaletsky H (1999) Primer3 on the WWW for general users and for biologist programmers. In: Bioinformatics methods and protocols. Springer, New York, pp 365–386
Schenkel FS, Miller SP, Ye X, Moore SS, Nkrumah JD, Li C, Yu J, Mandell IB, Wilton JW, Williams JL (2005) Association of single nucleotide polymorphisms in the leptin gene with carcass and meat quality traits of beef cattle. J Anim Sci 83(9):2009–2020
Schneider S, Kueffer J, Roessli D, Excoffier L (1997) Analysis of molecular variance inferred from metric distances among DNA haplotypes-application to human mitochondrial-DNA restriction data. Genetics 131:479–491
Singh R, Ong-Abdullah M, Low E-TL, Manaf MAA, Rosli R, Nookiah R, Ooi L-C, Ooi S-E, Chan K-L, Halim MA, Azizi N, Nagappan J, Bacher B, Lakey N, Smith SW, He D, Hogan M, Budiman MA, Lee EK, DeSalle R, Kudrna D, Goicoechea JL, Wing RA, Wilson RK, Fulton RS, Ordway JM, Martienssen RA, Sambanthamurthi R (2013) Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 500(7462):335–339
Stewart C, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14(5):748–750
Tang B, Tang M, Chen C, Qiu P, Liu Q, Wang M, Li C (2006) Characteristics of soil fauna community in the Dongjiao coconut plantation ecosystem in Hainan, China. Acta Ecol Sin 26(1):26–32
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106(3):411–422
Thurston M, Field D (2005) Msatfinder: detection and characterisation of microsatellites. Distributed by the authors at http://www.genomics.ceh.ac.uk/msatfinder/ CEH Oxford, Mansfield Road, Oxford OX1 3SR
Varshney RK, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett 7:537–546
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23(1):48–55
Wang M, Jiang N, Jia T, Leach L, Cockram J, Waugh R, Ramsay L, Thomas B, Luo Z (2012) Genome-wide association mapping of agronomic and morphologic traits in highly structured populations of barley cultivars. Theor Appl Genet 124(2):233–246
Xiao Y, Chen L, Zou J, Tian E, Xia W, Meng J (2010) Development of a population for substantial new type Brassica napus diversified at both A/C genomes. Theor Appl Genet 121(6):1141–1150
Xiao Y, Luo Y, Yang Y, Fan H, Xia W, Mason AS, Zhao S, Sager R, Fei Q (2013) Development of microsatellite markers in Cocos nucifera and their application in evaluating the level of genetic diversity of Cocos nucifera. Plant Omics 6(3):193–200
Xu J, Ranc N, Muños S, Rolland S, Bouchet J-P, Desplat N, Le Paslier M-C, Liang Y, Brunel D, Causse M (2013) Phenotypic diversity and association mapping for fruit quality traits in cultivated tomato and related species. Theor Appl Genet 26:567–581
Xue Y, Warburton ML, Sawkins M, Zhang X, Setter T, Xu Y, Grudloyma P, Gethi J, Ribaut J-M, Li W, Zhang X, Zheng Y, Yan J (2013) Genome-wide association analysis for nine agronomic traits in maize under well-watered and water-stressed conditions. Theor Appl Genet 126:2587–2596
Zhao Y, Williams R, Prakash CS, He G (2012) Identification and characterization of gene-based SSR markers in date palm (Phoenix dactylifera L.). BMC Plant Biol 12(1):237
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31101179), The Major Technology Project of Hainan (ZDZX2013023-1) and an Australian Research Council Discovery Early Career Researcher Award (DE120100668).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Wei Xia and Yong Xiao made equal contributions to the manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xia, W., Xiao, Y., Liu, Z. et al. Development of gene-based simple sequence repeat markers for association analysis in Cocos nucifera . Mol Breeding 34, 525–535 (2014). https://doi.org/10.1007/s11032-014-0055-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-014-0055-x