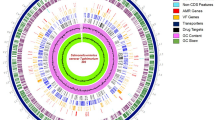
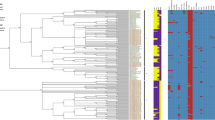
Abstract
In the era of emerging antibiotic resistance, Salmonella enterica subsp. enterica serovar Typhi the causative agent of typhoid, is a threat for healthcare systems in developing countries especially India, where the disease is highly endemic. Genetic diversity among different strains may be the cause of variable severity of disease in different regions of the world. To explore this genetic diversity, genome annotation by rapid annotation using subsystem technology (RAST) was carried out for genomes of four Salmonella Typhi strains from two distinct areas available in the public domain. Two clinical strains were from India (P-stx-12 and E02-1180) and the other two strains considered as reference strains were from the endemic regions of Papua New Guinea (UJ308A and UJ816A). We report that Indian clinical strains possess several similar genes responsible for virulence and pathogenicity as those present in the reference strains. Interestingly, Indian clinical strains also possess 34 additional potential virulence genes that are absent in the reference strains, suggesting the more dreadful nature of Indian clinical strains as compared to those causing endemic typhoid. Indian strains contained genes coding for; Colicin V and bacteriocin production; multidrug resistance efflux pumps; ABC transporters; Type III and Type VI secretion systems, siderophore aerobactin, pathogenicity islands and Vi polysaccharide biosynthesis and transport. These unique genes are also reported in the genomes of other six clinical strains of India analyzed through RAST and IslandViewer 4 for validation purpose. This study highlights the presence of potential genes as molecular targets to overcome the future endemic outbreaks in India.












Similar content being viewed by others
Abbreviations
- ABC:
-
ATP-binding cassette
- APEC:
-
Avian pathogenic E. coli
- hvKP:
-
Hypervirulent Klebsiella pneumoniae
- MDR:
-
Multidrug resistance
- PNG:
-
Papua New Guinea
- RAST:
-
Rapid annotation using subsystem technology
- SPI:
-
Salmonella pathogenicity island
- TTSS:
-
Type three secretion system
- WGS:
-
Whole genome sequencing
- WHO:
-
World Health Organization
- XDR:
-
Extensively drug resistant
References
Crump JA, Luby SP, Mintz ED (2004) The global burden of typhoid fever. Bull World Health Organ 82:346–353
Ochiai RL, Acosta CJ, Danovaro-Holliday MC, Baiqing D, Bhattacharya SK, Agtini MD et al (2008) A study of typhoid fever in five Asian countries: disease burden and implications for controls. Bull World Health Organ 86:260–268
Buckle GC, Walker CL, Black RE (2012) Typhoid fever and paratyphoid fever: systematic review to estimate global morbidity and mortality for 2010. J Glob Health 2:010401
Mogasale V, Maskery B, Ochiai RL, Lee JS, Mogasale VV, Ramani E et al (2014) Burden of typhoid fever in low-income and middle-income countries: a systematic, literature-based update with risk-factor adjustment. Lancet Glob Health 2:e570–e580
Lee JS, Mogasale VV, Mogasale V, Lee K (2016) Geographical distribution of typhoid risk factors in low and middle income countries. BMC Infect Dis 16:732
Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, Feasey NA et al (2015) Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet 47:632–639
Pham Thanh D, Thompson CN, Rabaa MA, Sona S, Sopheary S, Kumar V et al (2016) The molecular and spatial epidemiology of typhoid fever in rural Cambodia. PLoS Negl Trop Dis 10:e0004785
Techasaensiri C, Radhakrishnan A, Als D, Thisyakorn U (2018) Typhoidal Salmonella trends in Thailand. Am J Trop Med Hyg 99:64–71
Kavai SM, Kangogo M, Muigai AW, Kariuki S (2018) Analysis of trends in resistance to fluoroquinolones and extended spectrum beta-lactams among Salmonella Typhi isolates obtained from patients at four outpatient clinics in Nairobi County, Kenya. Adv Microbiol 8:578
Eng S-K, Pusparajah P, Ab Mutalib N-S, Ser H-L, Chan K-G, Lee L-H (2015) Salmonella: a review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci 8:284–293
Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK et al (2018) Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. MBio 9:e00105-18
Organization WH (2017) WHO publishes list of bacteria for which new antibiotics are urgently needed. WHO, Geneva
Zou Q-H, Li R-Q, Liu G-R, Liu S-L (2014) Comparative genomic analysis between typhoidal and non-typhoidal Salmonella serovars reveals typhoid-specific protein families. Infect Genet Evol 26:295–302
Hughes D, Andersson DI (2017) Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol Rev 41:374–391
Chan K, Baker S, Kim CC, Detweiler CS, Dougan G, Falkow S (2003) Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar typhimurium DNA microarray. J Bacteriol 185:553–563
McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A et al (2004) Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet 36:1268–1274
Divyashree S, Nabarro LE, Veeraraghavan B, Rupali P (2016) Enteric fever in India: current scenario and future directions. Trop Med Int Health 21:1255–1262
Kanungo S, Dutta S, Sur D (2008) Epidemiology of typhoid and paratyphoid fever in India. J Infect Dev Ctries 2:454–460
Banerjee T, Shukla BN, Filgona J, Anupurba S, Sen MR (2014) Trends of typhoid fever seropositivity over ten years in north India. Indian J Med Res 140:310–313
Chander AM, Nair RG, Kaur G, Kochhar R, Dhawan DK, Bhadada SK et al (2017) Genome insight and comparative pathogenomic analysis of Nesterenkonia jeotgali strain CD08_7 isolated from duodenal mucosa of celiac disease patient. Front Microbiol 8:129
Chander AM, Kochhar R, Dhawan DK, Bhadada SK, Mayilraj S (2018) Genome sequence and comparative genomic analysis of a clinically important strain CD11-4 of Janibacter melonis isolated from celiac disease patient. Gut Pathog 10:2
Kaur G, Chander AM, Kaur G, Maurya SK, Nadeem S, Kochhar R et al (2019) A genomic analysis of Mycobacterium immunogenum strain CD11_6 and its potential role in the activation of T cells against Mycobacterium tuberculosis. BMC Microbiol 19:64
Chander AM, Kumari M, Kochhar R, Dhawan DK, Bhadada SK, Mayilraj S (2017) Genome sequence of Kocuria polaris strain CD08_4, an isolate from the duodenal mucosa of a celiac disease patient. Genome Announc 5:e01158-17
Baddam R, Kumar N, Shaik S, Lankapalli AK, Ahmed N (2014) Genome dynamics and evolution of Salmonella Typhi strains from the typhoid-endemic zones. Sci Rep 4:7457
Baddam R, Thong KL, Avasthi TS, Shaik S, Yap KP, Teh CS et al (2012) Whole-genome sequences and comparative genomics of Salmonella enterica serovar Typhi isolates from patients with fatal and nonfatal typhoid fever in Papua New Guinea. J Bacteriol 194:5122–5123
Ong SY, Pratap CB, Wan X, Hou S, Abdul Rahman AY, Saito JA et al (2012) Complete genome sequence of Salmonella enterica subsp. enterica serovar Typhi P-stx-12. J Bacteriol 194:2115–2116
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genom 9:75
Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ et al (2015) RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T et al (2014) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214
Bertelli C, Laird MR, Williams KP, Simon Fraser University Research Computing G, Lau BY, Hoad G et al (2017) IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res 45:30–35
Saha R, Saha N, Donofrio RS, Bestervelt LL (2013) Microbial siderophores: a mini review. J Basic Microbiol 53:303–317
Chowdhury R, Sahu GK, Das J (1996) Stress response in pathogenic bacteria. J Biosci 21:149–160
Soto SM (2013) Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 4:223–229
Coyne S, Courvalin P, Perichon B (2011) Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother 55:947–953
Nagakubo S, Nishino K, Hirata T, Yamaguchi A (2002) The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J Bacteriol 184:4161–4167
Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J et al (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465
Springer B, Kidan YG, Prammananan T, Ellrott K, Bottger EC, Sander P (2001) Mechanisms of streptomycin resistance: selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob Agents Chemother 45:2877–2884
Jack RW, Tagg JR, Ray B (1995) Bacteriocins of gram-positive bacteria. Microbiol Rev 59:171–200
Riley MA, Wertz JE (2002) Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol 56:117–137
Yang SC, Lin CH, Sung CT, Fang JY (2014) Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front Microbiol 5:241
Gérard F, Pradel N, Wu L-F (2005) Bactericidal activity of colicin V is mediated by an inner membrane protein, SdaC, of Escherichia coli. J Bacteriol 187:1945–1950
Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubes R, Postle K et al (2007) Colicin biology. Microbiol Mol Biol Rev 71:158–229
Waters VL, Crosa JH (1991) Colicin V virulence plasmids. Microbiol Rev 55:437–450
Šmajs D, Micenková L, Šmarda J, Vrba M, Ševčíková A, Vališová Z et al (2010) Bacteriocin synthesis in uropathogenic and commensal Escherichia coli: colicin E1 is a potential virulence factor. BMC Microbiol 10:288
Pattus F, Massotte D, Wilmsen H, Lakey J, Tsernoglou D, Tucker A et al (1990) Colicins: prokaryotic killer-pores. Experientia Basel 46:180–192
Arai M, Liu L, Fujimoto T, Setiawan A, Kobayashi M (2011) DedA protein relates to action-mechanism of halicyclamine A, a marine spongean macrocyclic alkaloid, as an anti-dormant mycobacterial substance. Mar Drugs 9:984–993
Stegh AH, Schickling O, Ehret A, Scaffidi C, Peterhansel C, Hofmann TG et al (1998) DEDD, a novel death effector domain-containing protein, targeted to the nucleolus. EMBO J 17:5974–5986
Garmory HS, Titball RW (2004) ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun 72:6757–6763
Ge Y, Lee JH, Hu B, Zhao YF (2018) Loss-of-function mutations in the Dpp and Opp permeases render Erwinia amylovora resistant to kasugamycin and blasticidin S. Mol Plant Microbe Interact 31(8):823–832
Vander Broek CW, Stevens JM (2017) Type III secretion in the melioidosis pathogen Burkholderia pseudomallei. Front Cell Infect Microbiol 7:255
Lin J, Cheng J, Chen K, Guo C, Zhang W, Yang X et al (2015) The icmF3 locus is involved in multiple adaptation-and virulence-related characteristics in Pseudomonas aeruginosa PAO1. Front Cell Infect Microbiol 5:70
Kline KA, Dodson KW, Caparon MG, Hultgren SJ (2010) A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol 18:224–232
Ramos-Morales F (2012) Impact of Salmonella enterica type III secretion system effectors on the eukaryotic host cell. ISRN Cell Biol. https://doi.org/10.5402/2012/787934
Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H (1997) Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol 23:1089–1097
Hueck CJ (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62:379–433
Brown NF, Finlay BB (2011) Potential origins and horizontal transfer of type III secretion systems and effectors. Mob Genet Elements 1:118–121
Ehrbar K, Hardt WD (2005) Bacteriophage-encoded type III effectors in Salmonella enterica subspecies 1 serovar Typhimurium. Infect Genet Evol 5:1–9
Mirold S, Rabsch W, Tschape H, Hardt WD (2001) Transfer of the Salmonella type III effector sopE between unrelated phage families. J Mol Biol 312:7–16
Juhas M, Crook DW, Hood DW (2008) Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol 10:2377–2386
Christie PJ, Cascales E (2005) Structural and dynamic properties of bacterial type IV secretion systems (review). Mol Membr Biol 22:51–61
Ling J, Pan H, Gao Q, Xiong L, Zhou Y, Zhang D et al (2013) Aerobactin synthesis genes iucA and iucC contribute to the pathogenicity of avian pathogenic Escherichia coli O2 strain E058. PLoS ONE 8:e57794
Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA (2015) Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun 83:3325–3333
J-i Wachino, Arakawa Y (2012) Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist Updates 15:133–148
Macmaster R, Zelinskaya N, Savic M, Rankin CR, Conn GL (2010) Structural insights into the function of aminoglycoside-resistance A1408 16S rRNA methyltransferases from antibiotic-producing and human pathogenic bacteria. Nucleic Acids Res 38:7791–7799
Aguirre AA, Vicente AM, Hardwick SW, Alvelos DM, Mazzon RR, Luisi BF et al (2017) Association of the cold shock DEAD-box RNA helicase RhlE to the RNA degradosome in Caulobacter crescentus. J Bacteriol 199:e00135-00117
Netterling S, Bareclev C, Vaitkevicius K, Johansson J (2016) RNA helicase important for Listeria monocytogenes hemolytic activity and virulence factor expression. Infect Immun 84:67–76
Wetter M, Coulding D, Pickard D, Kowarik M, Waechter CJ, Dougan G, Wacker M (2012) Molecular characterization of the viaB locus encoding the biosynthetic machinery for Vi capsule formation in Salmonella Typhi. PLoS ONE 7:e45609
Liston SD, Ovchinnikova OG, Whitfield C (2016) Unique lipid anchor attaches Vi antigen capsule to the surface of Salmonella enterica serovar Typhi. Proc Natl Acad Sci USA A113:6719–6724
Tran QT, Gomez G, Khare S, Lawhon SD, Raffatellu M, Baumler AJ, Ajithdoss D, Dhavalas S, Adams LG (2010) The Salmonella enterica serovar Typhi Vi capsular antigen is expressed after the bacterium enters the ileal mucosa. Infect Immun 78:527–535
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors have approved the final draft of the manuscript and declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sekhon, P.K., Chander, A.M., Mayilraj, S. et al. Genomic analysis of Indian strains of Salmonella enterica subsp. enterica serovar Typhi indicates novel genetic repertoire for pathogenicity and adaptations. Mol Biol Rep 46, 3967–3989 (2019). https://doi.org/10.1007/s11033-019-04843-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04843-2