Abstract
Breeding for resistant crops is a sustainable way to control disease and relies on the introduction of novel resistance genes. Here, we tested three strategies on how to use transgenes from wheat to achieve durable resistance against fungal pathogens in the field. First, we tested the highly effective, overexpressed single transgene Pm3e in the background of spring wheat cultivar Bobwhite in a long-term field trial over many years. Together with previous results, this revealed that transgenic wheat line Pm3e#2 conferred complete powdery mildew resistance during a total of nine field seasons without a negative impact on yield. Furthermore, overexpressed Pm3e provided resistance to powdery mildew isolates from our worldwide collection when crossed into the elite wheat cultivar Fiorina. Second, we pyramided the four overexpressed transgenes Pm3a, Pm3b, Pm3d, and Pm3f in the background of cultivar Bobwhite and showed that the pyramided line Pm3a,b,d,f was completely resistant to powdery mildew in five field seasons. Third, we performed field trials with three barley lines expressing adult plant resistance gene Lr34 from wheat during three field seasons. Line GLP8 expressed Lr34 under control of the pathogen-inducible Hv-Ger4c promoter and provided partial barley powdery mildew and leaf rust resistance in the field with small, negative effects on yield components which might need compensatory breeding. Overall, our study demonstrates and discusses three successful strategies for achieving fungal disease resistance of wheat and barley in the field using transgenes from wheat. These strategies might confer long-term resistance if applied in a sustainable way.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Crop production is threatened by numerous pathogens leading to disease and yield losses in agriculture. Two approaches are commonly employed to combat diseases: the use of pesticides and the use of resistant crop cultivars. Breeding for resistant crops is the more sustainable approach and often relies on the identification and introduction of new resistance genes from the crop’s breeding pool. Pm3 is a major resistance gene conferring resistance to powdery mildew (Blumeria graminis f.sp. tritici, Bgt) in wheat. To date, there are 17 identified Pm3 alleles which provide resistance to different spectra of Bgt isolates (Yahiaoui et al. 2004, 2006, 2009; Srichumpa et al. 2005; Bhullar et al. 2009, 2010). They encode nucleotide-binding domain, leucine-rich repeat receptor proteins (NLRs) which confer race-specific resistance upon recognition of specific Bgt avirulence proteins (Avrs). To study and compare field resistance conferred by individual Pm3 alleles, transgenic wheat lines carrying single overexpressed Pm3 alleles were earlier created by biolistic transformation of spring wheat cultivar Bobwhite (Brunner et al. 2011, 2012; Koller et al. 2019). The allele, Pm3e specifically, was shown to confer strong powdery mildew resistance without any fitness cost, making it a potentially valuable allele for wheat breeding (Koller et al. 2019). However, race-specific resistance genes such as NLRs are prone to breakdown due to homogenous, large-scale deployment driving strong selection for gain-of-virulence pathogen mutations (Papaïx et al. 2018). An approach for improving resistance durability, as well as broadening the resistance spectrum, is the pyramiding of multiple resistance genes in a crop cultivar (Koller et al. 2018): There, for a successful infection, the pathogen must adapt multiple avirulence effectors to avoid recognition by the different resistance gene products. Pyramiding can be achieved using classical crossbreeding supported by marker-assisted approaches as was shown for the pyramiding of different Magnaporthe oryzae resistance genes in rice (Khan et al. 2018; Wu et al. 2019). However, pyramiding can also be achieved faster and more efficiently using genetic transformation approaches. In potato cultivar Desiree, for example, three late blight resistance genes were stacked using Agrobacterium tumefaciens-mediated transformation and the pyramided lines showed improved resistance compared to plants with single resistance genes during two field trials (Haesaert et al. 2015). Koller et al. (2018) described another example where transgenic wheat lines with two pyramided Pm3 alleles, generated by biolistic transformation and crossbreeding, showed enhanced powdery mildew resistance in the field compared to single transgenic lines. The improved resistance was attributed to additive transgene expression levels. Pyramiding of alleles of the same gene in a genetically stable way, as done for Pm3, can currently only be achieved by transformation and not by conventional crossing because the alleles are in the same genetic locus. Another class of resistance genes are quantitative, partially acting resistance genes. They are also referred to as adult plant resistance (APR) genes since resistance is only functional in the adult plant stage and not at the seedling stage as is the case for major resistance genes. Unlike major, race-specific resistance genes, APR genes do not completely abolish disease, rather they provide partial resistance which is characterized by reduced fungal growth. Consequently, they exert lower selection pressure on the pathogen population to adapt and overcome resistance and are generally considered to be more durable (Dinglasan et al. 2022). APR genes mostly confer partial and durable resistance to all races of a pathogen and in a few cases even to multiple pathogens (Kou and Wang 2010). Three genes conferring such broad-spectrum, multi-pathogen resistance were found in wheat. Lr34 (= Yr18/Sr57/Pm38), Lr46 (= Yr29/Sr58/Pm39), and Lr67 (= Yr46/Sr55/Pm46) confer partial resistance against all races of the fungal pathogens causing leaf rust (Puccinia triticina), stripe rust (Puccinia striiformis f.sp. tritici), stem rust (Puccinia graminis f.sp. tritici), and powdery mildew (Blumeria graminis f.sp. tritici) (Ellis et al. 2014; Krattinger et al. 2019). These genes are associated with leaf tip necrosis (LTN), a senescence-like phenotype in the flag leaf of adult plants (Singh 1992; Krattinger et al. 2016). Lr34 has become one of the most frequently used disease-resistance genes in wheat breeding and despite its widespread use has remained effective for over 100 years (Kolmer et al. 2008; Krattinger et al. 2013). Two predominant Lr34 alleles are found in wheat, but resistance is provided by only one (Lr34res), which evolved from the ancestral susceptible allele (Lr34sus) by two gain-of-function mutations (Krattinger et al. 2013). Lr34 encodes an ATP-binding cassette (ABC) transporter protein shown to mediate the transport of the phytohormone abscisic acid (ABA), which accumulates at the leaf tip (Krattinger et al. 2009, 2019; Bräunlich et al. 2021). However, the exact mechanisms of how Lr34 provides resistance remain elusive. Lr34 has been successfully transferred to most major cereals such as barley, rice, maize, durum wheat, and sorghum (Risk et al. 2013; Boni et al. 2018; Krattinger et al. 2016; Sucher et al. 2017; Rinaldo et al. 2017; Schnippenkoetter et al. 2017), indicating that the molecular mechanism for resistance must be conserved in all these plant species. In barley, Lr34 expression led to disease resistance already at the seedling stage, however, accompanied by premature LTN and reduced plant fitness (Risk et al. 2013; Chauhan et al. 2015). These negative effects were reduced without compromising resistance by using a pathogen-inducible promoter which decreased Lr34 expression levels in transgenic barley lines (Boni et al. 2018). In this study, we tested three approaches for improving field resistance to various biotrophic fungal pathogens using transgenes from wheat. First, we completed a long-term field study for powdery mildew resistance conferred by the overexpressed transgene Pm3e. Second, we tested pyramided line Pm3a,b,d,f overexpressing four transgenes during five field seasons. Third, we tested three barley lines expressing the adult plant resistance gene Lr34 from wheat during three field seasons for resistance to biotrophic fungal pathogens.
Materials and methods
Plant material
The wheat single transgenic lines Pm3a, Pm3b, Pm3d, and Pm3f in the background of spring wheat cultivar Bobwhite SH 98 26 were previously generated by Brunner et al. (2011, 2012) by biolistic transformation (named Pm3a#1, Pm3b#1, Pm3d#1, Pm3f#1). Lines Pm3a,d and Pm3b,f were previously generated and described by Stirnweis et al. (2014). Pyramided line Pm3a,b,d,f was obtained by crossing parental lines Pm3a,d and Pm3b,f, followed by several rounds of self-fertilization. The T5 generation was then analyzed by PCR for the presence of the four alleles Pm3a, Pm3b, Pm3d, and Pm3f using allele-specific primers. Pm3a primers: 5′-TGTATCATCTGGAACCAGCGT-3′ (forward); 5′-CCATAGTTGGATCAACTTCGCTA-3′ (reverse). Pm3b primers: 5′-TTTAGCCCTGCCTTCATACG-3′ (f); 5′-AGTAGCTCGGGAATCTTTCCA-3′ (r). Pm3d primers: 5′-GAATCCCTTTGGCTTGAAAGA-3′ (f); 5′-GCATAATCTGGAACATCGTATGG-3′ (r). Pm3f primers: 5′-CGGGTATTATTCCAGCACATGT-3′ (f); 5′-GAGTAGAAATGATTCTGTGCCTCTA-3′ (r). Based on this, the pyramided line homozygous for all four Pm3 alleles was selected. Transgenic Bobwhite line Pm3e#2 was previously generated and described by Koller et al. (2019). Pm3e-Fiorina BC3F2 seedlings were obtained by crossing Pm3e#2 and elite spring wheat cultivar Fiorina, followed by three back-crosses with Fiorina and subsequent selfing. Genotyping and anti-HA western blots confirmed the presence and expression of Pm3e-HA in each generation. The transgenic barley lines used in this study were BG9 generated and described in Risk et al. (2013), as well as GLP8 and GLP11 generated and described in Boni et al. (2018). In BG9, Lr34 expression is controlled by the native wheat promoter, while in GLP8 and GLP11, Lr34 expression is controlled by the pathogen-inducible promoter Hv-Ger4c. For seedling infection tests, the plants were grown in the greenhouse under long-day conditions (20/16 °C, 16 h photoperiod, 60% humidity).
Field trial set-up and disease scoring
Legal permits for field experiments involving genetically modified plants were obtained by the Federal Office for the Environment prior to the field trials (permits B18001, B18004, and B20002) under the Release Ordinance 2008 and the Gene Technology Act 2003 in compliance with the EU Directive 2001/18/EC. The field trials were performed during the five field seasons 2019, 2020, 2021, 2022, and 2023, essentially as described in Koller et al. (2019). Wheat and barley lines for resistance tests were grown in test plots of 1.5 m × 1.0 m at the so-called “Protected Site” (www.protectedsite.ch), an experimental field site for research trials with transgenic crops located at Agroscope in Zurich Reckenholz (Brunner et al. 2021; Romeis et al. 2013). Four test plots per genotype were grown in a randomized complete block design. Wheat powdery mildew test plots were flanked by infection rows consisting of the powdery mildew susceptible wheat breeding line FAL94632 and cultivar Kanzler. Pots with susceptible wheat plants pre-infected in the greenhouse with Swiss powdery mildew isolate Bgt 96224 (Wicker et al. 2013) were planted into the infection rows as described by Koller et al. (2019). Barley powdery mildew test plots were flanked by infection rows consisting of the powdery mildew susceptible barley cultivar Golden Promise inoculated with Blumeria graminis f.sp. hordei (Bgh) isolate K1 (Boni et al. 2018). Barley leaf rust test plots were artificially infected with Puccinia hordei isolate 1.2.1 (Risk et al. 2013). Scoring of powdery mildew and flowering was performed as described by Brunner et al. (2011, 2012). For leaf rust scoring, the percentage of flag leaf area covered with rust pustules was estimated approximately every 3 days after the onset of the disease. Yield plots were sown using a 7-row plot drill with 0.18-m row spacing and had a size of 6.5 m × 1.5 m. They were fungicide-treated. A sowing error in field season 2023 led to some replicate yield plots with less than the standard seven sowing rows. However, the number of seed sowed per plot was the same. Yield and 1000-grain weight were measured by five independent yield plot replicates per line and year.
Transgene expression analysis
Flag leaf samples (wheat) and penultimate leaf samples (barley) were collected from field-grown plants each year. For each of the four biological replicates (= four plots), three plant samples were pooled and frozen immediately in dry ice. RNA extraction was performed as described in Bräunlich et al. (2021) using the Dynabeads® mRNA DIRECT™ Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA). RNA (3 µL) was reverse transcribed in a total volume of 10 µL using the Maxima H Minus First Strand cDNA Synthesis Kit with dsDNase (Thermo Fisher Scientific). Quantitative reverse transcription PCR (RT-qPCR) experiments were performed according to MIQE guidelines (Bustin et al. 2009) using the KAPA SYBR® FAST qPCR Master Mix (Kapa Biosystems) on a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Reactions were run in technical triplicates and in 10 µL reaction volume using 4 µL of 1:10 diluted cDNA and 250 nM primers. For total Pm3 expression analysis in wheat, the following primers were used: 5′-CTGGAGTGTCTGTCGGGAGAG-3′ (forward) and 5′-GCATCTAGCCATTTGCGTTTG-3′ (reverse). These Pm3 primers did not distinguish between the four Pm3 alleles Pm3a, Pm3b, Pm3d, and Pm3f (Yahiaoui et al. 2004, 2006) since it was not possible to design reliable allele-specific qPCR primers due to high sequence similarity between them. Allele-specific primers previously designed for genotyping were tested for qPCR; however, they were not suitable. mRNA expression levels were normalized to reference gene Ta.6863 (Hurni et al. 2013; Koller et al. 2018, 2019). For Lr34 expression analysis in barley, the following primers were used: 5′-GACAGCGCCAGAATGGTGC-3′ (forward) and 5′-GACATCAACCCTGTCAATTC-3′ (reverse). ADP ribosylation factor (ADPRF) was used as a reference gene (Giménez et al. 2011). Gene expression data was analyzed using the CFX Maestro software version 2.2 (Bio-Rad).
Pm3-HA protein detection
Total protein was extracted from field-grown wheat flag leaf samples (three pooled leaves per plot) in protein extraction buffer containing 15 mM NaCl, 5 mM Tris–HCl pH 7.5, 0.5% Triton X-100 and one tablet cOmplete™ EDTA-free protease inhibitor cocktail (Roche). The total protein concentration of the extract was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Equal amounts of total protein were loaded and separated on 8% SDS polyacrylamide gels and transferred to a PVDF membrane. Anti-HA-HRP antibody (rat monoclonal, clone 3F10, Roche) was used in a 1:1000 dilution for detection of Pm3-HA, together with WesternBright EC HRP substrate (Advansta, San Jose, CA, USA). Chemiluminescence was captured using the Fusion FX Imaging System (Vilber Lourmat, Eberhardzell, Germany).
Powdery mildew infection tests at the seedling stage
Virulence phenotyping experiments at the seedling stage were performed by placing 3-cm-long first leaf segments of approximately 12-day-old wheat plants with the adaxial side up on 0.5% agar plates supplemented with 0.24 mM benzimidazole (Parlange et al. 2011). Leaf segments were infected with Bgt spores by homogenous spraying with a single-use glass pipette and incubated at 20 °C for 7 days with 16 h of light per day to allow for sufficient colony growth. Virulence scoring was performed 7 days after infection by estimating the percentage of leaf coverage with fungal colonies (LC). The virulence phenotype was then categorized into three classes: virulent for LC = 70–100%, intermediate for LC = 10–70%, and avirulent for LC > 10%. The powdery mildew-susceptible cultivar Kanzler was included as a control.
Statistical analyses
Data were presented as single data points and boxplots showing the median, the first, and the third quartile. Analysis of variance (ANOVA) followed by Tukey’s honestly significant difference (HSD) test was performed to evaluate statistical differences between groups using the “agricolae” package for RStudio version 4.1.3 (de Mendiburu 2021). Student’s t-tests were also carried out in RStudio. The significance threshold alpha = 0.05 was used for all analyses.
Results
Overexpressed Pm3e conferred strong powdery mildew field resistance during nine field seasons without negative impact on yield
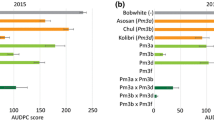
We tested transgenic Bobwhite event Pm3e#2, which overexpresses C-terminally HA-tagged Pm3e under the maize ubi promoter (Koller et al. 2019), during the five field seasons 2019 to 2023 for powdery mildew resistance. During the four field seasons from 2020 to 2023, the infection rows planted next to the test plots were artificially infected with Swiss powdery mildew isolate Bgt 96224 to ensure an evenly distributed powdery mildew infection in the field trial. In field season 2019, the powdery mildew infection originated entirely from the naturally occurring local powdery mildew population, which reliably causes powdery mildew disease every year at the field trial site in Zürich Reckenholz, Switzerland. Pm3e#2 showed strong powdery mildew resistance in all five field seasons 2019, 2020, 2021, 2022, and 2023 tested in this study (Fig. 1A). Together with our previous studies performed during field seasons 2015, 2016, 2017, and 2018 published in Koller et al. (2019), this resulted in a long-term study of nine field seasons in which Pm3e#2 showed strong powdery mildew resistance. In the four field seasons of 2019, 2021, 2022, and 2023 tested in this study, Pm3e#2 had AUDPC values = 0. The small powdery mildew infection in field season 2020 (AUDPC value average = 7) possibly originates from some seed admixtures during sowing (seed carryover from neighboring plots). In contrast, non-transformed Bobwhite and sister line Pm3e#2-sis (null segregant of Pm3e#2) were both powdery mildew susceptible in all tested field seasons (Pm3e#2-sis was only tested during field seasons 2021, 2022, and 2023) with AUDPC values up to 150 and 125, respectively (Fig. 1A). During the field seasons 2021 to 2023, we additionally tested if overexpressed Pm3e in transgenic event Pm3e#2 had a negative effect on yield compared to sister line Pm3e#2-sis and non-transformed Bobwhite. To test the yield potential in the absence of powdery mildew infection, we treated all yield plots with fungicides. In all three field seasons, there was no statistically significant difference in yield between Pm3e#2, Pm3e#2-sis, and non-transformed Bobwhite (Fig. 1B). However, the average amount of yield varied considerably between the different field seasons with an average of 3.2 kg per plot in 2021, 4.5 kg per plot in 2022, and 6.4 kg per plot in 2023. This could be explained by the prevailing weather conditions during the different field seasons, which can influence flowering time and duration of the grain-filling period, thereby influencing yield (Akter and Rafiqul Islam 2017; Yadav and Ellis 2017). In field season 2021, weather was generally unfavorable for wheat growth and wheat lines flowered later in June compared to the other field seasons (Suppl. Fig. S1A), likely leading to a reduced grain-filling period and generally lower yields. In addition to the yield data, we measured the 1000-grain weight of Pm3e#2, Pm3e#2-sis, and Bobwhite in field season 2023 and found no significant difference between the three lines (Fig. 1C).
Transgenic Pm3e confers strong powdery mildew resistance in the field. A Powdery mildew infection of field-grown Pm3e-transgenic wheat. The area under disease progress curve (AUDPC) values of four independent plots per genotype (except 2019: two plots per genotype) are shown. Differing letters indicate a significant difference in infection calculated by Tukey’s honestly significant difference test (Tukey HSD test, α = 0.050). Plot yield B and 1000-grain weight of field-grown Pm3e#2 C, its corresponding sister line Pm3e#-sis and untransformed Bobwhite. Yield and 1000-grain were measured from five independent, fungicide-treated plots per genotype. The letters above bars indicate a significant difference in yield or 1000-grain weight as determined by the Tukey HSD test (α = 0.050). D Representative pictures of Blumeria graminis f.sp. tritici (Bgt) infection tests with Fiorina (ubi::Pm3e) BC3F2 plants using two different Bgt isolates virulent on Fiorina and avirulent on Pm3e#2. For each isolate, 18 individual seedlings of Fiorina (ubi::Pm3e) BC3F2 were scored, and three biological replicates were scored for Fiorina, Pm3e#2, and Kanzler. As expected, there was segregation for the Pm3e transgene in the backcross genotypes. The susceptible wheat cultivar Kanzler was included as a positive control
Pm3e conferred powdery mildew resistance in elite wheat cultivar Fiorina
Furthermore, we sought to determine whether overexpressed Pm3e could be used to improve elite wheat cultivars with only intermediate powdery mildew resistance such as the spring wheat cultivar Fiorina (Strebel et al. 2022). We therefore performed a cross between Pm3e#2 and Fiorina, followed by three back-crosses with Fiorina, and tested powdery mildew resistance of the resulting, individual BC3F2 plants in seedling infection assays. We used Bgt isolates from our group’s worldwide Bgt collection and selected one Bgt isolate from Switzerland (Bgt CHE_96224), two from China (Bgt CHN_36_3 and Bgt CHN_36_70), and two from Israel (Bgt ISR_70 and Bgt ISR_103) which were virulent on parental cultivar Fiorina but avirulent on parental line Pm3e#2. We performed infection tests with the five Bgt isolates on Pm3e-Fiorina BC3F2—seedlings. The seedlings segregated in the expected 3:1 ratio for powdery mildew resistance: susceptibility over all five tested Bgt isolates, showing the expected powdery mildew resistance effect of Pm3e in Pm3e-Fiorina (results for Bgt CHN_36_70 and Bgt IRS_70 are shown in Fig. 1D; X2 (1, N = 18) = 0.67, p = 0.41, for both isolates). Together, these results showed that overexpressed Pm3e provided strong powdery mildew resistance during nine field seasons without compromising yield and demonstrated its potential to improve powdery mildew resistance in elite wheat cultivars.
Pyramiding of four overexpressed Pm3 alleles in wheat line Pm3a,b,d,f resulted in strong powdery mildew resistance during five field seasons
We generated the transgenic pyramided wheat line Pm3a,b,d,f in the background of cultivar Bobwhite by crossing parental transgenic Bobwhite lines Pm3a,d and Pm3b,f, which carry the two overexpressed transgenes Pm3a and Pm3d, and the two overexpressed transgenes Pm3b and Pm3f, respectively. We self-fertilized the progeny of the crosses for up to five generations and selected the pyramided line which carries all four transgenes in a homozygous state. All transgenes were expressed under the maize ubiquitin (ubi) promoter and were C-terminally fused to the sequence encoding the HA epitope tag for protein detection, except for Pm3b, which has no epitope tag, i.e., ubi::Pm3a-HA, ubi::Pm3b, ubi::Pm3d-HA, and ubi::Pm3f-HA. We then tested pyramided line Pm3a,b,d,f during five field seasons for powdery mildew resistance. During all five field seasons, pyramided wheat line Pm3a,b,d,f showed complete powdery mildew resistance with AUDPC values = 0 (Fig. 2A). In contrast, the powdery mildew-susceptible, non-transformed Bobwhite had AUDPC values ranging between 50 and 150, depending on the field season’s weather-dependent disease pressure. In field seasons 2021, 2022, and 2023, we also grew the two parental lines Pm3a,d and Pm3b,f, and the four grand parental lines Pm3a, Pm3b, Pm3d, and Pm3f as additional controls. Each grand parental line carries one of the four overexpressed transgenes Pm3a, Pm3b, Pm3d, or Pm3f. Consistent with previous studies, the grand parental line Pm3f was also completely powdery mildew resistant with AUDPC values = 0 in all three field seasons (Fig. 2A). Similarly, the grand parental line Pm3b showed very strong powdery mildew resistance in all three tested field seasons, while lines Pm3a and Pm3d showed only intermediate resistance but with clearly less symptoms than susceptible Bobwhite. We collected flag leaf samples of the field-grown transgenic wheat lines to measure total Pm3 expression and total Pm3-HA protein accumulation. RT-qPCR analysis showed higher total Pm3 expression in line Pm3a,b,d,f compared to the grand parental lines in 2021, 2022, and 2023 (Fig. 2B). Expression data from 2023 suggests additive expression levels of the four Pm3 alleles in pyramided line Pm3a,b,d,f; however, data from 2021 and 2022 did not confirm truly additive expression levels of the four Pm3 alleles (Fig. 2B). Similarly inconclusive results were obtained from the protein analysis: total Pm3-HA protein accumulation data of field-grown flag leaf samples did not show any obvious additive Pm3-HA accumulation in line Pm3a,b,d,f. Surprisingly, the total amount of Pm3-HA protein accumulation in wheat line Pm3a,b,d,f was similar to the amount of Pm3f-HA protein accumulation in grand parental line Pm3f (Fig. 2C).
Pyramided wheat line Pm3a,b,d,f shows strong powdery mildew resistance in the field. A Powdery mildew infection of field-grown, Pm3 allele-pyramided transgenic wheat. The area under disease progress curve (AUDPC) values of four independent plots per genotype (except for 2019: five plots Pm3a,b,d,f and two plots non-transformed Bobwhite) are shown. Differing letters indicate significant differences in infection (Tukey HSD test, α = 0.050). B Total Pm3 expression relative to reference gene Ta.6863 in field-grown flag leaf samples determined by RT-qPCR. Letters above bars indicate significant differences in expression levels (Tukey HSD test, α = 0.050). C Total Pm3-HA protein accumulation in flag leaf samples from field plants. Each sample contains three pooled flag leaf segments from different plant individuals. Total protein concentration was measured and adjusted to the same concentration prior to loading. Ponceau staining indicates equal loading
To evaluate possible pleiotropic effects of the overexpressed transgenes in the transgenic lines, we additionally scored flowering dates of the field-grown lines. In field season 2021, there was no significant difference in flowering time between pyramided line Pm3a,b,d,f and untransformed Bobwhite; however, in field seasons 2022 and 2023, Pm3a,b,d,f had a slight delay in the flowering of approximately 2 days, similar to Pm3f (Suppl. Fig. S1A). Chlorotic leaves were observed in field-grown grand parental line Pm3f as described previously by Brunner et al. (2012). Interestingly, in pyramided line Pm3a,b,d,f, this pleiotropic effect was significantly diminished (Suppl. Fig. S1B). Overall, these results showed that pyramiding of the four Pm3 alleles Pm3a, Pm3b, Pm3d, and Pm3f provided complete powdery mildew resistance during five field seasons while reducing the leaf chlorosis observed in the single transgenic line Pm3f.
Wheat transgene Lr34 provides leaf rust and powdery mildew resistance in field-grown transgenic barley
We grew three transgenic barley lines expressing the wheat transgene Lr34 in field seasons 2021, 2022, and 2023 and tested them for barley leaf rust (Puccinia hordei) and barley powdery mildew (Blumeria graminis f. sp. hordei (Bgh)) resistance. Barley line BG9 expresses Lr34 under the native Lr34 promoter from wheat (Risk et al. 2013), while barley lines GLP8 and GLP11 express Lr34 under the pathogen-inducible promoter Hv-Ger4c, which originates from a barley gene (Boni et al. 2018). Barley leaf rust resistance test plots were artificially infected with P. hordei isolate 1.2.1. In the two field seasons 2021 and 2022, all three transgenic barley lines BG9, GLP8, and GLP11 were significantly more resistant against barley leaf rust compared to their corresponding sister lines BG9-sis, GLP8-sis, GLP11-sis, and untransformed barley background cultivar Golden Promise (GP) (Fig. 3A). In field season 2023, however, GLP8 showed a similar level of rust infection as GLP8-sis and GP, possibly because differences might not have been clearly visible due to the lower disease pressure, reflected by the reduced overall AUDPC values in 2023 compared to 2022 and 2021.
Lr34 from wheat provides leaf rust and powdery mildew resistance in field-grown transgenic barley lines BG9, GLP8, and GLP11. Leaf rust infection (A) and powdery mildew infection (B). The area under disease progress curve (AUDPC) values of four independent plots per genotype are shown. C Flowering dates of field-grown barley lines. Differing letters indicate significant differences in infection or flowering time (Tukey HSD test, α = 0.050)
Barley powdery mildew resistance test plots were artificially infected with Bgh isolate K1. In all tested field seasons, all three transgenic barley lines BG9, GLP8, and GLP11 were significantly more resistant against barley powdery mildew compared to their corresponding sister lines, as well as compared to untransformed GP (Fig. 3B). Furthermore, we scored flowering dates of all field-grown barley lines to detect possible pleiotropic effects of the Lr34 transgene. In all three field seasons, there was no difference in flowering time between the transgenic- and sister lines, except for GLP11 which flowered significantly later than all other lines (Fig. 3C). In field season 2023, we additionally grew GLP8 and GLP11 together with their corresponding sister lines and GP in fungicide treated yield plots. GLP8 showed only a slight reduction in yield compared to GLP8-sis and GP; however, GLP11 showed a significant reduction in yield (Suppl. Fig. S2A). Likewise, the 1000-grain weight was very similar among the tested lines, except for GLP11 which had a significantly reduced 1000-grain weight (Suppl. Fig. S2B). This coincides with the delayed flowering of GLP11, which likely led to a shortened grain-filling period and therefore reduced yield. Another pleiotropic effect observed in the field was leaf tip necrosis (LTN) visible at the adult stage in BG9 but not in any of the other transgenic lines (Fig. 4A). Moreover, in field season 2023, we collected penultimate leaf samples from all field-grown barley lines to determine the expression of the Lr34 transgene by an RT-qPCR assay. Line BG9 showed by far the highest Lr34 expression of all transgenic lines, with approximately 100-fold higher expression levels than GLP8 and GLP11 (Fig. 4B). This coincided with previous studies describing stronger Lr34 expression when expressed under the native wheat promoter than under the pathogen-inducible Hv-Ger4c promoter under greenhouse conditions (Boni et al. 2018) and under field conditions (Bräunlich et al. 2021). From all these observations, we concluded that Lr34 provided resistance to two different pathogens, Bgh and P. hordei, in barley during three field seasons, however, with varying strength and sometimes pleiotropic effects. Importantly, transgenic barley line GLP8 showed partial resistance to both pathogens without any negative pleiotropic effects.
Leaf tip necrosis (LTN) and RT-qPCR assay in field-grown barley of field season 2023. A Photograph of barley penultimate leaves. For each line, two leaves from two different plant individuals are shown. B Lr34 expression relative to reference gene ADP ribosylation factor (ADPRF) in field-grown barley penultimate leaves. The asterisks (*) indicate p < 0.05 calculated by Student’s t-test
Discussion
High breeding value of Pm3e
Transgenic line Pm3e#2 showed complete powdery mildew resistance during five field seasons (Fig. 1A). Conversely, the corresponding sister line Pm3e#2-sis and untransformed Bobwhite were completely powdery mildew susceptible. Thus, resistance was only provided by the Pm3e transgene. This complements previously conducted field trials, where Pm3e#2 already provided strong powdery mildew resistance during four field seasons (2015–2018) (Koller et al. 2019). Taken together, these data over 9 years from consecutive field seasons demonstrate that overexpressed Pm3e confers strong and long-term powdery mildew resistance in the field. Even though no breakdown of Pm3e-provided resistance was observed during nine field seasons, it is not unlikely to happen in the future if deployed at large-scale, as has been shown for other major resistance genes such as Pm8 (Bennett 1984; Kunz et al. 2023) and Sr31 (Pretorius et al. 2000; Singh et al. 2011). Therefore, combining Pm3e with other resistance genes to increase durability must be considered for the sustainable use of this promising allele in wheat breeding. Furthermore, our study showed no negative impact of overexpressed Pm3e on yield potential or 1000-grain weight (Fig. 1B and C), showing that wheat cultivar Bobwhite is a suitable background for Pm3e transgene expression. However, Bobwhite is an agronomically rather uninteresting cultivar, so we tested the possibility of introducing Pm3e into an elite wheat cultivar to improve its powdery mildew resistance. We crossed Pm3e into elite spring wheat cultivar Fiorina, which only shows intermediate powdery mildew resistance, and showed that Pm3e-Fiorina BC3F2—seedlings were completely resistant to various powdery mildew isolates virulent on Fiorina (Fig. 1D). Our seedlings still segregated for Pm3e, and more rounds of self-fertilization and genotyping would be needed to obtain stably homozygous Pm3e-Fiorina plants for seed multiplication and subsequent field trials. Nonetheless, our results demonstrated the successful broad-spectrum resistance action of Pm3e in the background of elite cultivar Fiorina. Compatibility with the genetic background is essential for correct transgene function and the absence of pleiotropic effects. This has, for instance, been demonstrated in pepper, where efficiency of the major resistance genes Me1 and Me3, conferring resistance to nematodes, was affected by the genetic background into which they were introduced (Barbary et al. 2014). Another example was shown for the late blight resistance gene RB, which was more efficient when introduced into potato cultivar Kufri Jyoti than in cultivar Kufri Bahar (Shandil et al. 2017). Alongside the field results presented in this study, Pm3e shows high potential in providing strong powdery mildew resistance without any negative pleiotropic effects, making it a promising allele for (elite) wheat breeding programs.
Benefits and limitations of pyramiding four overexpressed Pm3 alleles
Pyramiding of resistance genes has frequently been used in breeding to improve durability and increase the spectrum of resistance. Koller et al. (2018) showed that pyramiding of two Pm3 alleles resulted in strong powdery mildew resistance in the field. Here, we showed that combining the four Pm3 alleles Pm3a, Pm3b, Pm3d, and Pm3f resulted in complete powdery mildew resistance during five field seasons (Fig. 2A). Transgene expression analyses further revealed a tendency towards higher total Pm3 expression in pyramided line Pm3a,b,d,f compared to the single transgenic lines (Fig. 2B). Since the primers used for our expression analysis did not distinguish between the different Pm3 alleles, we cannot infer how much each Pm3 allele contributed to the total Pm3 expression in pyramided line Pm3a,b,d,f. However, genotyping confirmed the presence of all four Pm3 alleles in Pm3a,b,d,f. Complete powdery mildew field resistance and similar protein accumulation of both Pm3a,b,d,f and Pm3f could suggest that only allele Pm3f is active and conferring resistance in the pyramided line, or that Pm3f is the dominant allele fully explaining the phenotype. Our expression data, however, showed higher, although not significantly, total Pm3 expression in line Pm3a,b,d,f compared to line Pm3f. Importantly, the reduction of the Pm3f chlorotic leaf phenotype in the pyramided line suggests that the other Pm3 alleles contribute to the phenotype and are not simply silenced. Transgene silencing and suppression are phenomena which have been observed before when combining different transgenes or alleles and should be considered when breeding pyramided lines (Rajput et al. 2022; Zorrilla-López et al. 2013; Hurni et al. 2014; Stirnweis et al. 2014; Koller et al. 2023). For resistance gene Pm3 particularly, functional suppression among different Pm3 alleles was previously shown in seedling infection tests when combined in double Pm3-transgenic wheat lines (Stirnweis et al. 2014). The suppression effect, which occurred at the post-translational level, depended on the combination of the two Pm3 alleles and on the specific powdery mildew isolate that was used for infection. However, in field trials with the double Pm3-transgenic lines, two of which are the parental lines of our pyramided line Pm3a,b,d,f, no suppression was observed (Koller et al. 2018). We observed an example of transgene silencing when pyramiding powdery mildew resistance genes Pm17 and Pm3CS in the same background of wheat cultivar Bobwhite. In this case, ubi promoter silencing likely led to the transcriptional silencing of both transgenes and thus powdery mildew susceptibility of the pyramided line (Koller et al. 2023). While it has been suggested that using the same promoter repeatedly can induce homology-dependent gene silencing, there are also many reports of transgenic plants containing several transgenes under the same promoter without any silencing effects (Zorrilla-López et al. 2013; Koller et al. 2023). In the previously mentioned field trials, Koller et al. (2018) attributed the enhanced powdery mildew resistance of the double Pm3-transgenic lines compared to the single transgenic lines to additive transgene expression and allele-specific combinations. We thus speculate that this also applies to pyramided line Pm3a,b,d,f, broadening the resistance spectrum and possibly increasing the durability of resistance. Furthermore, pyramiding diminished the pleiotropic phenotype of line Pm3f, restoring plant development in Pm3a,b,d,f to that of untransformed Bobwhite. Altogether, a pyramiding of four overexpressed Pm3 alleles proved to be a successful approach for providing complete powdery mildew resistance in the field during five field seasons. To determine if this resistance is durable, field studies in many different environments as well as over long time, ultimately in a released variety, would be needed (Johnson 1984). Given the current situation for GMO plant breeding and growth in Europe, this is not feasible and can only be realized if regulations would be simplified.
Fine-tuning of Lr34 expression allows for disease resistance with reduced pleiotropic effects in barley
Previously, Lr34-transgenic barley lines BG9 and GLP8 were tested in a small trial during one field season (Bräunlich et al. 2021). In that study, BG9 showed complete resistance to barley powdery mildew and barley leaf rust as opposed to its sister line, however, accompanied by a strong LTN phenotype. We confirmed this complete resistance phenotype of BG9 during three additional field seasons and also observed strong LTN (Figs. 3A, B and 4A). Line GLP8 showed partial resistance to both pathogens in our field trials, except in field season 2023, where the same level of leaf rust disease as its corresponding sister line was observed. Further, we showed that Lr34 in transgenic line GLP11 provided almost complete resistance to P. hordei and partial resistance to Bgh during three field seasons. Neither GLP11 nor GLP8 displayed LTN in the field. As demonstrated previously in greenhouse-grown wheat and barley, strong Lr34 expression is associated with LTN and correlates with upregulation of senescence marker genes and several pathogenesis-related (PR) genes (Krattinger et al. 2009; Risk et al. 2012, 2013; Chauhan et al. 2015). Consistently, our expression analysis showed the strongest Lr34 expression in line BG9 (Fig. 4B). While upregulation of PR genes in Lr34-expressing wheat occurs after rust infection (Hulbert et al. 2007), PR gene upregulation in transgenic barley expressing Lr34 under the native wheat promoter is constitutive (Chauhan et al. 2015). The accumulation of abscisic acid (ABA) at the leaf tip in Lr34-barley seems to be involved in the induction of PR genes (Bräunlich et al. 2021). In contrast to wheat, Lr34 under the native wheat promoter is constitutively expressed in barley already at the seedling stage leading to an early LTN phenotype and reduced yield components as was shown for line BG9 in the greenhouse (Risk et al. 2013). To reduce the pleiotropic effects of native Lr34 expression in barley, lines GLP8 and GLP11 were generated which express Lr34 under the pathogen-inducible promoter Hv-Ger4c at a lower level than BG9 (Boni et al. 2018). Indeed, lines GLP8 and GLP11 showed higher total grain weight compared to BG9 under greenhouse conditions; however, under near-field conditions, GLP11 showed reduced yield parameters. When tested under field conditions, line BG9 performed rather poorly with visible LTN and significantly reduced 1000-grain weight (Bräunlich et al. 2021); hence, we did not measure yield parameters for BG9 in our field trial. We showed that the 1000-grain weight of GLP11 but not GLP8 was significantly reduced under field conditions compared to the untransformed cultivar Golden Promise (Suppl. Fig. S2B). Since sister line GLP11-sis (null segregant) showed no reduction, we conclude that this is most likely a pleiotropic effect of the Lr34 transgene in GLP11 and not any tissue culture effect. The total yield of line GLP8 was similar to that of its sister line GLP8-sis in the field, but line GLP11 showed a slight reduction compared to GLP11-sis. This is in accordance with previous results obtained under greenhouse conditions (Boni et al. 2018). Since the pleiotropic effects of Lr34 were found to be expression level-dependent in barley (Chauhan et al. 2015), it is crucial to fine-tune Lr34 expression to reduce such negative effects while still maintaining strong disease resistance. Line GLP8 demonstrated a successful first attempt at using a pathogen-inducible promoter leading to lower Lr34 expression, no LTN, improved yield parameters, and nonetheless partial resistance to P. hordei and Bgh in the field. Similarly, a Lr34-expressing rice transgenic line showed lower Lr34 expression and reduced LTN compared to other transgenic lines, yet still showed partial resistance to the rice blast fungus Magnaporthe oryzae (Krattinger et al. 2016). Moreover, it was also shown in Lr34-expressing durum wheat that LTN is not required for disease resistance (Rinaldo et al. 2017). Thus, it would be necessary to generate a large number of transgenic barley lines, using different and tissue-specific inducible promoters, to achieve optimal Lr34 expression, which still confers resistance but with no or minimal pleiotropic effects. This highlights the importance of analyzing and adjusting transgene expression levels, especially when transferring genes with large physiological effects such as Lr34 and Lr67 (Chauhan et al. 2015; Milne et al. 2019). It is also likely that in a breeding program with large genetic diversity, genetic backgrounds with some compensatory activity for the pleiotropic effects of Lr34 can be identified.
Conclusion
Importance of field trials and future considerations for breeding crops with durable disease resistance
Besides disease resistance, the agronomic performance in the field is of central interest for crop breeders. Therefore, it is indispensable to test newly generated lines also for plant fitness in the field setting and not only in the greenhouse since certain pleiotropic effects are only visible in the field. This was for example shown for two other transgenic wheat lines overexpressing Pm3b, which showed chlorotic leaves only when grown in the field but not in the greenhouse (Brunner et al. 2011), similar to line Pm3f used in this study. We observed a reduction of the chlorotic leaf phenotype in Pm3a,b,d,f, and we did not detect any pleiotropic effects negatively impacting yield in line Pm3e#2. In two Lr34-barley lines, we observed different pleiotropic effects, such as strong LTN in line BG9, delayed flowering, and lower yield in line GLP11. These negative effects, however, could most likely be reduced by fine-tuning expression levels of Lr34 to achieve disease resistance without pleiotropic effects as demonstrated in line GLP8.
The genetic backgrounds of the transgenic lines in this study were cultivars Bobwhite for wheat and Golden Promise for barley, chosen based on their high transformation efficiency (Pellegrineschi et al. 2002; Schreiber et al. 2020). However, easily transformable plant genotypes often do not perform as well agronomically as elite cultivars and so are of little interest to breeders and farmers. Therefore, promising resistance genes tested successfully in the field, such as Pm3e and Lr34 in this study, could be introduced into elite wheat and barley cultivars. We demonstrated a promising first attempt by introducing Pm3e into elite wheat cultivar Fiorina, which provided resistance to powdery mildew isolates which were virulent on Fiorina. Recent advances in genome editing technologies could facilitate the targeted introgression of resistance genes into different cultivars and speed up the lengthy breeding process (Pixley et al. 2022). For example, a new method called HI-Edit enables delivering the CRISPR-Cas9 genome-editing machinery components more efficiently into elite crop germplasm (Kelliher et al. 2019). Further, CRISPR-Cas9 has also been adapted for multiplexed editing, i.e., the simultaneous targeting of multiple DNA loci (Wang and Doudna 2023). This is particularly relevant for polyploid crops like wheat, which carry many redundant alleles or copies of the target gene. To take advantage of all the technological progress for improved crop breeding, however, genome-edited plants would need to become more politically and publicly accepted.
Data availability
All data supporting the findings of this study are included in the paper and its supplementary data or are available from the corresponding author on request.
References
Akter N, Rafiqul Islam M (2017) Heat stress effects and management in wheat A review. Agron Sustain Dev 37(5):1–17. https://doi.org/10.1007/S13593-017-0443-9
Barbary A, Palloix A, Fazari A, Marteu N, Castagnone-Sereno P, Djian-Caporalino C (2014) The plant genetic background affects the efficiency of the pepper major nematode resistance genes Me1 and Me3. Theor Appl Genet 127(2):499–507. https://doi.org/10.1007/s00122-013-2235-1
Bennett FGA (1984) Resistance to powdery mildew in wheat: a review of its use in agriculture and breeding programmes. Plant Pathol 33(3):279–300. https://doi.org/10.1111/J.1365-3059.1984.TB01324.X
Bhullar NK, Street K, Mackay M, Yahiaoui N, Keller B (2009) Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc Natl Acad Sci USA 106(23):9519–9524. https://doi.org/10.1073/pnas.0904152106
Bhullar NK, Zhang Z, Wicker T, Keller B (2010) Wheat gene bank accessions as a source of new alleles of the powdery mildew resistance gene Pm3: a large scale allele mining project. BMC Plant Biol 10(1):1–13. https://doi.org/10.1186/1471-2229-10-88
Boni R, Chauhan H, Hensel G, Roulin A, Sucher J, Kumlehn J, Brunner S, Krattinger SG, Keller B (2018) Pathogen-inducible Ta-Lr34res expression in heterologous barley confers disease resistance without negative pleiotropic effects. Plant Biotechnol J 16(1):245–253. https://doi.org/10.1111/PBI.12765
Bräunlich S, Koller T, Glauser G, Krattinger SG, Keller B (2021) Expression of the wheat disease resistance gene Lr34 in transgenic barley leads to accumulation of abscisic acid at the leaf tip. Plant Physiol Biochem 166:950–957. https://doi.org/10.1016/j.plaphy.2021.07.001
Brunner S, Hurni S, Herren G, Kalinina O, von Burg S, Zeller SL, Schmid B, Winzeler M, Keller B (2011) Transgenic Pm3b wheat lines show resistance to powdery mildew in the field. Plant Biotechnol J 9(8):897–910. https://doi.org/10.1111/J.1467-7652.2011.00603.X
Brunner S, Stirnweis D, Diaz Quijano C, Buesing G, Herren G, Parlange F, Barret P, Tassy C, Sautter C, Winzeler M, Keller B (2012) Transgenic Pm3 multilines of wheat show increased powdery mildew resistance in the field. Plant Biotechnol J 10(4):398–409. https://doi.org/10.1111/J.1467-7652.2011.00670.X
Brunner S, Romeis J, Patocchi A, Peter Agroscope R (2021) The protected site-seven years of field research with genetically modified plants. Agrarforschung Schweiz 12:9–15. https://doi.org/10.34776/afs12-9e
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4):611–622. https://doi.org/10.1373/CLINCHEM.2008.112797
Chauhan H, Boni R, Bucher R, Kuhn B, Buchmann G, Sucher J, Selter LL, Hensel G, Kumlehn J, Bigler L, Glauser G, Wicker T, Krattinger SG, Keller B (2015) The wheat resistance gene Lr34 results in the constitutive induction of multiple defense pathways in transgenic barley. Plant J 84(1):202–215. https://doi.org/10.1111/tpj.13001
Dinglasan E, Periyannan S, Hickey LT (2022) Harnessing adult-plant resistance genes to deploy durable disease resistance in crops. Essays Biochem 66(5):571–580. https://doi.org/10.1042/EBC20210096
Ellis JG, Lagudah ES, Spielmeyer W, Dodds PN (2014) The past, present and future of breeding rust resistant wheat. Front Plant Sci 5(641) https://doi.org/10.3389/fpls.2014.00641
Giménez MJ, Pistón F, Atienza SG (2011) Identification of suitable reference genes for normalization of qPCR data in comparative transcriptomics analyses in the Triticeae. Planta 233(1):163–173. https://doi.org/10.1007/s00425-010-1290-y
Haesaert G, Vossen JH, Custers R, De Loose M, Haverkort A, Heremans B, Hutten R, Kessel G, Landschoot S, Van Droogenbroeck B, Visser RGF, Gheysen G (2015) Transformation of the potato variety Desiree with single or multiple resistance genes increases resistance to late blight under field conditions. Crop Prot 77:163–175. https://doi.org/10.1016/J.CROPRO.2015.07.018
Hulbert SH, Bai J, Fellers JP, Pacheco MG, Bowden RL (2007) Gene expression patterns in near isogenic lines for wheat rust resistance gene Lr34/Yr18. Phytopathology 97(9):1083–1093. https://doi.org/10.1094/PHYTO-97-9-1083
Hurni S, Brunner S, Buchmann G, Herren G, Jordan T, Krukowski P, Wicker T, Yahiaoui N, Mago R, Keller B (2013) Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J 76(6):957–969. https://doi.org/10.1111/tpj.12345
Hurni S, Brunner S, Stirnweis D, Herren G, Peditto D, McIntosh RA, Keller B (2014) The powdery mildew resistance gene Pm8 derived from rye is suppressed by its wheat ortholog Pm3. Plant J 79(6):904–913. https://doi.org/10.1111/tpj.12593
Johnson R (1984) A Critical Analysis of Durable Resistance. Annu Rev Phytopathol 22(1):309–330. https://doi.org/10.1146/annurev.py.22.090184.001521
Kelliher T, Starr D, Su X, Tang G, Chen Z, Carter J, Wittich PE, Dong S, Green J, Burch E, McCuiston J, Gu W, Sun Y, Strebe T, Roberts J, Bate NJ, Que Q (2019) One-step genome editing of elite crop germplasm during haploid induction. Nature Biotechnol 37(3):287–292. https://doi.org/10.1038/s41587-019-0038-x
Khan GH, Shikari AB, Vaishnavi R, Najeeb S, Padder BA, Bhat ZA, Parray GA, Bhat MA, Kumar R, Singh NK (2018) Marker-assisted introgression of three dominant blast resistance genes into an aromatic rice cultivar Mushk Budji. Sci Rep 8(1):1–13. https://doi.org/10.1038/s41598-018-22246-4
Koller T, Brunner S, Herren G, Hurni S, Keller B (2018) Pyramiding of transgenic Pm3 alleles in wheat results in improved powdery mildew resistance in the field. Theor Appl Genet 131(4):861–871. https://doi.org/10.1007/s00122-017-3043-9
Koller T, Brunner S, Herren G, Sanchez-Martin J, Hurni S, Keller B (2019) Field grown transgenic Pm3e wheat lines show powdery mildew resistance and no fitness costs associated with high transgene expression. Transgenic Res 28(1):9–20. https://doi.org/10.1007/s11248-018-0099-5
Koller T, Camenzind M, Jung E, Brunner S, Herren G, Armbruster C, Keller B, Ch MC (2023) Pyramiding of transgenic immune receptors from primary and tertiary wheat gene pools improves powdery mildew resistance in the field. J Exp Bot. https://doi.org/10.1093/JXB/ERAD493
Kolmer JA, Singh RP, Garvin DF, Viccars L, William HM, Huerta-Espino J, Ogbonnaya FC, Raman H, Orford S, Bariana HS, Lagudah ES (2008) Analysis of the Lr34/Yr18 rust resistance region in wheat germplasm. Crop Sci 48(5):1841–1852. https://doi.org/10.2135/CROPSCI2007.08.0474
Kou Y, Wang S (2010) Broad-spectrum and durability: understanding of quantitative disease resistance. Curr Opin Plant Biol 13(2):181–185. https://doi.org/10.1016/J.PBI.2009.12.010
Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323(5919):1360–1363. https://doi.org/10.1126/science.1166453
Krattinger SG, Jordan DR, Mace ES, Raghavan C, Luo MC, Keller B, Lagudah ES (2013) Recent emergence of the wheat Lr34 multi-pathogen resistance: insights from haplotype analysis in wheat, rice, sorghum and Aegilops tauschii. Theor Appl Genet 126(3):663–672. https://doi.org/10.1007/s00122-012-2009-1
Krattinger SG, Sucher J, Selter LL, Chauhan H, Zhou B, Tang M, Upadhyaya NM, Mieulet D, Guiderdoni E, Weidenbach D, Schaffrath U, Lagudah ES, Keller B (2016) The wheat durable, multipathogen resistance gene Lr34 confers partial blast resistance in rice. Plant Biotechnol J 14(5):1261–1268. https://doi.org/10.1111/PBI.12491
Krattinger SG, Kang J, Bräunlich S, Boni R, Chauhan H, Selter LL, Robinson MD, Schmid MW, Wiederhold E, Hensel G, Kumlehn J, Sucher J, Martinoia E, Keller B (2019) Abscisic acid is a substrate of the ABC transporter encoded by the durable wheat disease resistance gene Lr34. New Phytol 223(2):853–866. https://doi.org/10.1111/NPH.15815
Kunz L, Sotiropoulos AG, Graf J, Razavi M, Keller B, Müller MC (2023) The broad use of the Pm8 resistance gene in wheat resulted in hypermutation of the AvrPm8 gene in the powdery mildew pathogen. BMC Biology 21(1):1–15. https://doi.org/10.1186/S12915-023-01513-5
de Mendiburu F (2021) agricolae: statistical procedures for agricultural research. R Package Version 1.3–5. https://CRAN.R-project.org/package=agricolae
Milne RJ, Dibley KE, Schnippenkoetter W, Mascher M, Lui ACW, Wang L, Lo C, Ashton AR, Ryan PR, Lagudah ES (2019) The wheat Lr67 gene from the sugar transport protein 13 family confers multipathogen resistance in barley. Plant Physiol 179(4):1285–1297. https://doi.org/10.1104/PP.18.00945
Papaïx J, Rimbaud L, Burdon JJ, Zhan J, Thrall PH (2018) Differential impact of landscape-scale strategies for crop cultivar deployment on disease dynamics, resistance durability and long-term evolutionary control. Evol Appl 11(5):705–717. https://doi.org/10.1111/EVA.12570
Parlange F, Oberhaensli S, Breen J, Platzer M, Taudien S, Šimková H, Wicker T, Doležel J, Keller B (2011) A major invasion of transposable elements accounts for the large size of the Blumeria graminis f.sp. tritici genome. Funct Integr Genomics 11(4):671–677. https://doi.org/10.1007/S10142-011-0240-5
Pellegrineschi A, Noguera LM, Skovmand B, Brito RM, Velazquez L, Salgado MM, Hernandez R, Warburton M, Hoisington D (2002) Identification of highly transformable wheat genotypes for mass production of fertile transgenic plants. Genome 45(2):421–430. https://doi.org/10.1139/G01-154
Pixley KV, Falck-Zepeda JB, Paarlberg RL, Phillips PWB, Slamet-Loedin IH, Dhugga KS, Campos H, Gutterson N (2022) Genome-edited crops for improved food security of smallholder farmers. Nature Genet 54(4):364–367. https://doi.org/10.1038/s41588-022-01046-7. Nature Research
Pretorius ZA, Singh RP, Wagoire WW, Payne TS (2000).Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis. f. sp. tritici in Uganda. Plant Dis 84(2) https://doi.org/10.1094/PDIS.2000.84.2.203B
Rajput R, Naik J, Misra P, Trivedi PK, Pandey A (2022) Gene pyramiding in transgenic plant development: approaches and challenges. J Plant Growth Regul 42(10):6038–6056. https://doi.org/10.1007/s00344-022-10760-9
Rinaldo A, Gilbert B, Boni R, Krattinger SG, Singh D, Park RF, Lagudah E, Ayliffe M (2017) The Lr34 adult plant rust resistance gene provides seedling resistance in durum wheat without senescence. Plant Biotechnol J 15(7):894–905. https://doi.org/10.1111/PBI.12684
Risk JM, Selter LL, Krattinger SG, Viccars LA, Richardson TM, Buesing G, Herren G, Lagudah ES, Keller B (2012) Functional variability of the Lr34 durable resistance gene in transgenic wheat. Plant Biotechnol J 10(4):477–487. https://doi.org/10.1111/J.1467-7652.2012.00683.X
Risk JM, Selter LL, Chauhan H, Krattinger SG, Kumlehn J, Hensel G, Viccars LA, Richardson TM, Buesing G, Troller A, Lagudah ES, Keller B (2013) The wheat Lr34 gene provides resistance against multiple fungal pathogens in barley. Plant Biotechnol J 11(7):847–854. https://doi.org/10.1111/PBI.12077
Romeis J, Meissle M, Brunner S, Tschamper D, Winzeler M (2013) Plant biotechnology: research behind fences. Trends Biotechnol 31(4):222–224. https://doi.org/10.1016/J.TIBTECH.2013.01.020
Schnippenkoetter W, Lo C, Liu G, Dibley K, Chan WL, White J, Milne R, Zwart A, Kwong E, Keller B, Godwin I, Krattinger SG, Lagudah E (2017) The wheat Lr34 multipathogen resistance gene confers resistance to anthracnose and rust in sorghum. Plant Biotechnol J 15(11):1387–1396. https://doi.org/10.1111/PBI.12723
Schreiber M, Mascher M, Wright J, Padmarasu S, Himmelbach A, Heavens D, Milne L, Clavijo BJ, Stein N, Waugh R (2020) A genome assembly of the barley ‘transformation reference’ cultivar golden promise. G3: Genes\Genomes\Genetics 10(6):1823. https://doi.org/10.1534/G3.119.401010
Shandil RK, Chakrabarti SK, Singh BP, Sharma S, Sundaresha S, Kaushik SK, Bhatt AK, Sharma NN (2017) Genotypic background of the recipient plant is crucial for conferring RB gene mediated late blight resistance in potato. BMC Genet 18(1):1–8. https://doi.org/10.1186/s12863-017-0490-x
Singh RP (1992) Expression of wheat leaf rust resistance gene Lr34 in seedlings and adult plants. Plant Dis 76(5):489. https://doi.org/10.1094/PD-76-0489
Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, Njau P, Herrera-Foessel S, Singh PK, Singh S, Govindan V (2011) The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol 49:465–481. https://doi.org/10.1146/ANNUREV-PHYTO-072910-095423
Srichumpa P, Brunner S, Keller B, Yahiaoui N (2005) Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol 139(2):885–895. https://doi.org/10.1104/PP.105.062406
Stirnweis D, Milani SD, Brunner S, Herren G, Buchmann G, Peditto D, Jordan T, Keller B (2014) Suppression among alleles encoding nucleotide-binding-leucine-rich repeat resistance proteins interferes with resistance in F1 hybrid and allele-pyramided wheat plants. Plant J 79(6):893–903. https://doi.org/10.1111/tpj.12592
Strebel S, Levy Häner L, Mattin M, Schaad N, Morisoli R, Watroba M, Girard M, Courvoisier N, Berberat J, Grandgirard R, Graf B, Streit M, Weisflog T (2022) Liste der empfohlenen Getreidesorten für die Ernte 2023(7) https://ira.agroscope.ch/de-CH/publication/49386
Sucher J, Boni R, Yang P, Rogowsky P, Büchner H, Kastner C, Kumlehn J, Krattinger SG, Keller B (2017) The durable wheat disease resistance gene Lr34 confers common rust and northern corn leaf blight resistance in maize. Plant Biotechnol J 15(4):489–496. https://doi.org/10.1111/PBI.12647
Wang JY, Doudna JA (2023) CRISPR technology: A decade of genome editing is only the beginning. Science 379(6629):eadd8643. https://doi.org/10.1126/science.add8643
Wicker T, Oberhaensli S, Parlange F, Buchmann JP, Shatalina M, Roffler S, Ben-David R, Doležel J, Šimková H, Schulze-Lefert P, Spanu PD, Bruggmann R, Amselem J, Quesneville H, Ver Loren Van Themaat E, Paape T, Shimizu KK, Keller B (2013) The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nature Genet 45(9):1092–1096. https://doi.org/10.1038/ng.2704
Wu Y, Xiao N, Chen Y, Yu L, Pan C, Li Y, Zhang X, Huang N, Ji H, Dai Z, Chen X, Li A (2019) Comprehensive evaluation of resistance effects of pyramiding lines with different broad-spectrum resistance genes against Magnaporthe oryzae in rice (Oryza sativa L.). Rice 12(1):1–13. https://doi.org/10.1186/S12284-019-0264-3
Yadav G, Ellis RH (2017) Effects of rain shelter or simulated rain during grain filling and maturation on subsequent wheat grain quality in the UK. J Agric Sci 155(2):300–316. https://doi.org/10.1017/S0021859616000411
Yahiaoui N, Srichumpa P, Dudler R, Keller B (2004) Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 37(4):528–538. https://doi.org/10.1046/J.1365-313X.2003.01977.X
Yahiaoui N, Brunner S, Keller B (2006) Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J 47(1):85–98. https://doi.org/10.1111/j.1365-313X.2006.02772.x
Yahiaoui N, Kaur N, Keller B (2009) Independent evolution of functional Pm3 resistance genes in wild tetraploid wheat and domesticated bread wheat. Plant J 57(5):846–856. https://doi.org/10.1111/J.1365-313X.2008.03731.X
Zorrilla-López U, Masip G, Arjó G, Bai C, Banakar R, Bassie L, Berman J, Farré G, Miralpeix B, Pérez-Massot E, Sabalza M, Sanahuja G, Vamvaka E, Twyman RM, Christou P, Zhu C, Capell T (2013) Engineering metabolic pathways in plants by multigene transformation. Int J Dev Biol 57(6-7–8):565–576. https://doi.org/10.1387/IJDB.130162PC
Acknowledgements
We thank the field technicians from Agroscope in Zurich–Reckenholz for their help with the field trial.
Funding
Open access funding provided by University of Zurich. Funding was provided by grant 310030_192526 from the Swiss National Science Foundation.
Author information
Authors and Affiliations
Contributions
TK, MC, EJ, and SB designed and carried out the field trials. MC, CA, TK, and GH performed the experiments. MC, TK, and BK wrote the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Camenzind, M., Koller, T., Armbruster, C. et al. Breeding for durable resistance against biotrophic fungal pathogens using transgenes from wheat. Mol Breeding 44, 8 (2024). https://doi.org/10.1007/s11032-024-01451-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-024-01451-2