Abstract
Key message
The combined effects of enhanced total transgene expression level and allele-specificity combination in transgenic allele-pyramided Pm3 wheat lines result in improved powdery mildew field resistance without negative pleiotropic effects.
Abstract
Allelic Pm3 resistance genes of wheat confer race-specific resistance to powdery mildew (Blumeria graminis f. sp. tritici, Bgt) and encode nucleotide-binding domain, leucine-rich repeat (NLR) receptors. Transgenic wheat lines overexpressing alleles Pm3a, b, c, d, f, and g have previously been generated by transformation of cultivar Bobwhite and tested in field trials, revealing varying degrees of powdery mildew resistance conferred by the transgenes. Here, we tested four transgenic lines each carrying two pyramided Pm3 alleles, which were generated by crossbreeding of lines transformed with single Pm3 alleles. All four allele-pyramided lines showed strongly improved powdery mildew resistance in the field compared to their parental lines. The improved resistance results from the two effects of enhanced total transgene expression levels and allele-specificity combinations. In contrast to leaf segment tests on greenhouse-grown seedlings, no allelic suppression was observed in the field. Plant development and yield scores of the pyramided lines were similar to the mean scores of the corresponding parental lines, and thus, the allele pyramiding did not cause any negative effects. On the contrary, in pyramided line, Pm3b × Pm3f normal plant development was restored compared to the delayed development and reduced seed set of parental line Pm3f. Allele-specific RT qPCR revealed additive transgene expression levels of the two Pm3 alleles in the pyramided lines. A positive correlation between total transgene expression level and powdery mildew field resistance was observed. In summary, allele pyramiding of Pm3 transgenes proved to be successful in enhancing powdery mildew field resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In agricultural practice, two important strategies to control crop diseases are the use of pesticides and the use of disease-resistant cultivars. To generate resistant cultivars, a common approach is the introgression of resistance genes found in the gene pool of the crop species of interest by multiple steps of backcrossing (Visscher et al. 1996; Hillel et al. 1990; Tanksley and Nelson 1996). A faster and more precise method of introgression is the genetic transformation of high yielding crop cultivars with resistance genes of interest (Narusaka et al. 2013; Rodriguez-Moreno et al. 2017; Lacombe et al. 2010; Mondal et al. 2016). Resistance gene Pm3 from wheat confers resistance to powdery mildew (Blumeria graminis f. sp. tritici, Bgt). So far, 17 functional Pm3 alleles have been identified (Yahiaoui et al. 2004, 2006; Srichumpa et al. 2005; Bhullar et al. 2009, 2010). The Pm3 alleles encode nucleotide-binding domain; leucine-rich repeat (NLR) type of receptors which are able to recognize effectors from Bgt and subsequently trigger a hypersensitive immune response. The interaction of wheat and Bgt is complex, involving many Bgt effectors, some of which act as avirulence factors as in case of effector AvrPm3a2f2 (Bourras et al. 2015). AvrPm3a2f2 has been identified as being recognized by the two Pm3 variants Pm3a and Pm3f (Bourras et al. 2015). Recently, effector SvrPm3a1f1, which acts as a suppressor of recognition, has been identified, disclosing another layer of complexity of the wheat–Bgt interaction (Bourras et al. 2015, 2016; Parlange et al. 2015). Wheat lines carrying Pm3 transgenes have been generated by biolistic transformation of spring wheat cultivar Bobwhite (Brunner et al. 2011, 2012) which does not carry an endogenous Pm3 allele. Four transgenic wheat lines carrying the allele Pm3b at different loci in the genome were generated and tested in field trials. The Pm3b lines were more powdery mildew resistant than their corresponding sister lines (null segregants of the transgene) (Brunner et al. 2011). In addition to the Pm3b lines, transgenic wheat lines carrying alleles Pm3a, Pm3c, Pm3d, Pm3f, and Pm3g were generated and two lines for alleles Pm3a, Pm3c, and Pm3f and one line for alleles Pm3d and Pm3g and all corresponding sister lines were tested in the field, together with three line mixtures consisting of 1:1 seed mixtures of lines Pm3a + Pm3b, Pm3a + Pm3d, and Pm3b + Pm3d. The transgenic lines showed increased powdery mildew resistance compared to their sister lines and the multilines were more powdery mildew resistant than the transgenic lines in pure stands (Brunner et al. 2012).
Several resistance genes or several alleles of the same resistance gene can be combined, or as often called “pyramided”, in one plant genotype to further improve resistance in terms of durability and spectrum. If several pyramided resistance genes are present in the plant encoding receptors which are able to detect different effectors of a pathogen, the pathogen has to modify multiple effectors to not trigger an immune response in the plant. If several pyramided alleles of a resistance gene are present in the same plant and these alleles encode receptors, which are able to detect different effectors present in different isolates of the pathogen population, the plant is able to detect a wider range of the pathogen population. Thus, pyramiding of resistance genes and resistance alleles has the potential to prevent or delay the development of boom-and-bust cycles commonly observed when only single- resistance genes are deployed (McDonald and Linde 2002; Zhan et al. 2015; Burdon et al. 2014; Delmotte et al. 2016). Pyramiding of resistance genes can be achieved by classical crossbreeding or by genetic engineering. Pyramiding of several alleles of the same resistance gene can only be achieved by genetic engineering. A prominent example of successful resistance gene pyramiding by genetic engineering is the combination of late blight-resistance genes in potato (Zhu et al. 2012; Jo et al. 2014). An example of gene pyramiding by genetic engineering for improved insect resistance is the combination of transgenes encoding Bacillus thuringiensis (Bt) toxins in maize and rice (reviewed in Liu et al. 2016). Stirnweis et al. (2014) generated Pm3 allele-pyramided lines by crossbreeding transgenic lines carrying single Pm3 alleles. They generated the four double homozygous lines Pm3a × Pm3b, Pm3a × Pm3d, Pm3b × Pm3d, and Pm3b × Pm3f. They tested the powdery mildew resistance of the pyramided lines in infection tests on leaf segments in the laboratory. Surprisingly, for some but not all Bgt isolates, a resistance suppression was observed, namely Pm3 variants Pm3b and Pm3c suppressed the function of the two Pm3 variants, Pm3a and Pm3f (Stirnweis et al. 2014). Pm3b suppressed the triggering of a hypersensitive response of autoactivated Pm3f in Nicotiana benthamiana assays and the authors were able to pinpoint the LRR domain as the domain conferring suppression (Stirnweis et al. 2014). Furthermore, in co-immunoprecipitation assays, the authors showed an interaction of Pm3b and Pm3f (Stirnweis et al. 2014). The objective of this study was to test the pyramided lines Pm3a × Pm3b, Pm3a × Pm3d, Pm3b × Pm3d, and Pm3b × Pm3f, all in the background of cultivar Bobwhite, in the field, to assess powdery mildew field resistance and pleiotropic effects of the pyramided transgenes on plant development and yield.
Materials and methods
Transgenic wheat lines
Transgenic wheat lines in the genetic background of spring wheat cultivar Bobwhite SH 98 26 carrying alleles Pm3a, b, d, or f and their corresponding sister lines were previously generated and described by Brunner et al. (2011, 2012) (named Pm3a#1, Pm3b#1, Pm3d#1, Pm3f#1, Sa#1, Sb#1, Sd#1, and Sf#1). Pyramided lines Pm3a × Pm3b, Pm3a × Pm3d, Pm3b × Pm3d, and Pm3b × Pm3f were previously generated and described by Stirnweis et al. (2014). The following seed generations were used in 2015 field trial: T4 (Pm3b, sister line Pm3f), T5 (Pm3a, Pm3d, Pm3f), F5 (Pm3a × Pm3d, Pm3b × Pm3d), and F6 (Pm3a × Pm3b, Pm3b × Pm3f). The following seed generations were used in 2016 field trial: T4 (sister line Pm3f), T5 (Pm3b), T6 (Pm3a, Pm3d, Pm3f), F6 (Pm3a × Pm3d, Pm3b × Pm3d), and F7 (Pm3a × Pm3b, Pm3b × Pm3f). Seeds of the transgenic lines and the corresponding sister line were produced in the field in 2014 and 2015, respectively.
Field trial
Field trials were carried out at the “Protected Site” of the Swiss centre of excellence for agricultural research Agroscope in Zurich—Reckenholz during field seasons 2015 and 2016. The “Protected Site” is a site for field trials with genetically modified plants (http://www.protectedsite.ch; Romeis et al. 2013). The site is protected to prevent vandalism. Microplots in both years had a size of 1.32 m × 1.0 m. Five plots per line or cultivar were grown in a randomized complete block design. In 2015, yield plots had a size of 6.0 m × 1.5 m and in 2016 a size of 5.8 m × 1.5 m. Four plots per line were grown in a randomized complete block design. We determined germination rates and thousand kernel weights of each line and cultivar and sowed 400 viable kernels per m2. Seeds were treated with 2 ml/kg Celest Trio (Syngenta) and 1.5 ml/kg Smaragd (Bayer). Each yield plot was bordered on all sides by plots of powdery mildew-resistant triticale cultivar Trado, leading to a chessboard-like design, and in addition, 1.32 m-wide transverse rows of triticale between the short sides of all neighboring plots to avoid direct corner-to-corner contact between the test plots. Micro and yield plots were flanked by infection rows consisting of the powdery mildew susceptible wheat breeding line FAL94632 and cultivar Kanzler. Pots with susceptible wheat plants pre-infected in the greenhouse with Bgt isolate 96224 were planted into the infection rows. The test area was surrounded by a 3 m-wide border crop consisting of triticale cultivar Trado. Application of fertilizer and insecticides was performed according to the standard Swiss agricultural practices. No fungicides were applied. Powdery mildew disease symptoms and plant development phenotypes were scored as previously described by Brunner et al. (2011, 2012).
Pm3 transgene expression analyses by RT qPCR
Leaf samples were collected in 2015 and 2016 from plants grown in microplots. Segments of the fully developed flag leaves from three plants per plot were pooled in one tube and immediately frozen on dry ice and stored at − 80 °C. Expression of Pm3 was quantified in a reverse transcription, quantitative real-time PCR (RT qPCR) assay, using a CFX384 Real-Time System C1000TM Thermal cycler (Bio-Rad). RNA extraction was performed as described by Hurni et al. (2015), and first-strand cDNA was synthesized from 0.5 μg RNA, using 1/2 reaction of the iScript Advanced cDNA Kit (172-5038, Bio-Rad). RT qPCR was performed with 2.4 µl of 20-fold-diluted cDNA in 6 µl and technical triplicates. Specificities of amplicons, RT-minus control check, and efficiency calculation were performed as described in Hurni et al. (2015). Relative quantities were calculated and normalized to the reference gene Ta.6863 revealing the calibrated normalized relative quantities (CNRQ) values, using the program qbase+ V 3.0 (Biogazelle).
Results
Field trials of pyramided lines and evaluation of the Bgt population present in the field
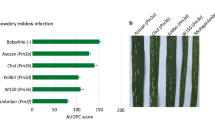
In the field trials, we tested the four pyramided lines Pm3a × Pm3b, Pm3a × Pm3d, Pm3b × Pm3d, and Pm3b × Pm3f, their parental lines Pm3a, Pm3b, Pm3d and Pm3f, and the sister lines of the parental lines at a field site for trials with genetically modified plants in Switzerland. The sister lines are null segregants of the transgene, meaning that no Pm3 alleles are present. We included in the field trial non-transformed Bobwhite and spring wheat cultivars Asosan, Chul, and Kolibri, which carry the endogenous alleles Pm3a, Pm3b, and Pm3d, respectively. Line mixtures Pm3a + Pm3b, Pm3a + Pm3d, Pm3b + Pm3d, and Pm3b + Pm3f were included in the yield trial for performance comparison of the pyramided line and the line mixture. To increase disease pressure, we planted spreader rows between each line of plots consisting of powdery mildew susceptible wheat plants infected with Bgt isolate 96224. This isolate was originally collected in Switzerland and since then cultivated in the laboratory. It was chosen for field infection, because previously conducted leaf segment tests in the laboratory showed avirulence of the isolate on wheat lines carrying any of the alleles Pm3a, Pm3b, Pm3d, or Pm3f (Brunner et al. 2010, 2011, 2012). We scored powdery mildew infection starting from the time point of onset of the disease, which occurred around 2 months after sowing and subsequently calculated the area under disease progress curve (AUDPC) score for each line. In 2015, Chul (Pm3b) (AUDPC score 205) was almost as highly infected as non-transformed Bobwhite (AUDPC score 233), Asosan (Pm3a) had an AUDPC score of 160, thus was slightly resistant, and Kolibri (Pm3d) with an AUDPC score of 83 was the least infected of the cultivars carrying an endogenous Pm3 allele (Fig. 1a). In 2016, disease severity was weaker compared to 2015 probably due to the weather conditions. However, the infection pattern in 2016 was very similar: non-transformed Bobwhite was the most infected cultivar with an AUDPC score of 188 followed by Asosan and Chul with similar scores of 136 and 142, respectively, and Kolibri with a score of 91 (Fig. 1b). In summary, in both years, cultivars Asosan (Pm3a) and Chul (Pm3b) were almost as infected as non-transformed Bobwhite, and cultivar Kolibri (Pm3d) showed a medium level of powdery mildew resistance (Fig. 1). Thus, in both years, Bgt strains virulent on alleles Pm3a, Pm3b, and Pm3d were present in the field. In 2015, we placed pots in the field containing winter wheat cultivar Michigan Amber/8*CC which contains the endogenous Pm3f allele. The potted plants showed powdery mildew infections (data not shown), and thus, Bgt strains virulent on Pm3f were present in the field as well.
Powdery mildew infection of field grown transgenic pyramided lines (dark green), transgenic parental lines (light green), cultivars carrying endogenous Pm3 alleles (yellow), and non-transformed Bobwhite (grey). Means of area under disease progress curve (AUDPC) scores were calculated from five independent replicates (five microplots) per line grown in field season 2015 (a) and in field season 2016 (b). Standard errors of five replicates are indicated (color figure online)
The four allele-pyramided lines are more powdery mildew resistant in the field than their parental lines carrying a single Pm3 allele
Line Pm3a had AUDPC scores of 179 and 99 in 2015 and 2016, respectively (Fig. 1a, b). Line Pm3d had similar AUDPC scores as line Pm3a in both years (Fig. 1a, b). Line Pm3b had AUDPC scores of 99 and 16 in 2015 and 2016, respectively (Fig. 1a, b). Line Pm3f was completely powdery mildew resistant in both years with AUDPC scores of 0 (Fig. 1a, b). Thus, the transgenic allele Pm3f provided the best, Pm3b an intermediate and alleles Pm3a and Pm3d the weakest disease resistance. In agreement with this observation, pyramided line Pm3a × Pm3d was the least powdery mildew resistant among the four pyramided lines in both years (Fig. 1a, b). In 2015, it had an AUDPC score of 104, which still was lower than the scores of their parental lines (Pm3d: AUDPC score 149 and Pm3a: AUDPC score 179) (Fig. 1a). In 2016, pyramided line Pm3a × Pm3d had an AUDPC score of 35, which as well was lower than the score of their parental lines (Pm3a: AUDPC score 99 and Pm3d: AUDPC score 104) (Fig. 1b). Pyramided line Pm3b × Pm3d had AUDPC scores of 22 in 2015 and 4 in 2016, respectively, and thus showed only trace infections and was more resistant than its parental lines Pm3b and Pm3d (Fig. 1a, b). The two pyramided lines Pm3a × Pm3b and Pm3b × Pm3f in both years were completely powdery mildew resistant with AUDPC scores of 0 (Fig. 1a, b). Taken together, these results show an improved powdery mildew resistance in the pyramided lines compared to their parental lines.
Agronomic traits of the pyramided lines are similar to the mean of the corresponding parental lines
To assess potential pleiotropic effects on plant development caused by pyramiding of Pm3 alleles, we scored flowering dates and seed set rates of the field grown lines. The summer 2015 was above-average sunny, warm, and dry, and overall, the plants developed well. In 2016, the weather conditions were not conducive to wheat growth. It was mostly rainy and moist, and the plants were stressed. Thus, negative effects on plant development were more distinct in 2016 compared to 2015; however, the overall plant development patterns were similar in both years (Fig. 2). In 2015, non-transformed Bobwhite, parental lines Pm3a, Pm3b, and Pm3d as well as sister line Pm3f and the pyramided lines Pm3a × Pm3b, Pm3a × Pm3d, and Pm3b × Pm3d all flowered within the same 2 days, whereas line Pm3f flowered almost 1 week later (Fig. 2a). Interestingly, pyramided line Pm3b × Pm3f only flowered 2 days later than non-transformed Bobwhite, and thus, the late flowering phenotype of line Pm3f was partly restored in the progeny line Pm3b × Pm3f (Fig. 2a). In 2015, line Pm3f showed a low seed set rate of 68% compared to the seed set rates of 89–95% of all the other lines, including pyramided line Pm3b × Pm3f (Fig. 2b). In 2016, non-transformed Bobwhite, parental line Pm3d, sister line Pm3f, and pyramided line Pm3a × Pm3d all flowered within the same 2 days (Fig. 2c). Pyramided line Pm3b × Pm3d flowered 2 days later than non-transformed Bobwhite and parental lines Pm3b and Pm3a flowered 3 and 4 days later than non-transformed Bobwhite, respectively (Fig. 2c). Pyramided line Pm3a × Pm3b flowered 4 days later than non-transformed Bobwhite as well (Fig. 2c). Parental line Pm3f flowered 9 days later than non-transformed Bobwhite (Fig. 2c). Again, this late flowering phenotype was partly restored in progeny pyramided line Pm3b × Pm3f, which flowered 7 days after non-transformed Bobwhite (Fig. 2c). In 2016, line Pm3f had a low seed set rate of 65% compared to all the other lines with seed set rates of 82–94% (Fig. 2d). The low seed set rate of Pm3f was largely restored in progeny line Pm3b × Pm3f with a rate of 82% (Fig. 2d).
Flowering date (a, c) and seed set rate (b, d) of pyramided lines (dark green), parental lines (light green), non-transformed Bobwhite (grey), and sister line Pm3f (grey) are shown for plants grown in field season 2015 (a, b) and 2016 (c, d). Standard errors of five replicates (five microplots per line) are indicated (color figure online)
Allele-pyramided lines are more powdery mildew resistant than the corresponding line mixtures in the yield plots; however, the pyramided lines and the corresponding line mixtures have similar yields
In 2016, in addition to the microplots, we grew all the parental lines, the pyramided lines, and the corresponding line mixtures in larger yield plots (Fig. 3a). Line mixtures consisted of a 1:1 ratio seed mixture of the parental lines in the following four combinations: Pm3a + Pm3b, Pm3a + Pm3d, Pm3b + Pm3d, and Pm3b + Pm3f. Overall, the powdery mildew AUDPC scores of the lines grown in the yield plots (Fig. 3b) were similar to the AUDPC score of the lines grown in the microplots (Fig. 1). The line mixtures had similar AUDPC scores compared to the mean of the AUDPC scores of the corresponding lines in pure stands (Fig. 3b). Compared to the corresponding pyramided lines, all four line mixtures had higher AUDPC scores, and thus, the pyramided lines were more powdery mildew resistant than the corresponding line mixtures (Fig. 3b). In terms of yield, there was no significant difference between the pyramided lines and the corresponding line mixtures (Fig. 4). Overall, the parental lines, the pyramided lines, and the line mixtures had similar yield, except for lines and mixtures containing Pm3f (Fig. 4). Parental line Pm3f had a significantly lower yield than all the other lines (Fig. 4). The severe yield loss phenotype of line Pm3f was partly restored in progeny pyramided line Pm3b × Pm3f, which had a similar yield as line mixture Pm3b + Pm3f (Fig. 4).
a Photograph of field trial 2016. The yield plots are arranged in a chessboard-like design, bordered on all sides by plots of powdery mildew-resistant triticale cultivar Trado. On the left border of the field, the three rows containing the smaller so-called microplots are visible. b Powdery mildew infection in yield plots of pyramided lines (dark green), line mixtures (yellow), components of line mixtures in pure stand/parental lines (light green), and non-transformed Bobwhite (grey). Mean of area under disease progress curve (AUDPC) scores was calculated of four independent replicates (four yield plots) per line grown in field season 2016 (color figure online)
Yields of pyramided lines, line mixtures, components of line mixtures in pure stand/parental lines and non-transformed Bobwhite grown in yield plots in 2016. Medians are indicated of four independent replicates (four yield plots) per line. Same letters indicate no significant difference in pairwise comparisons
Transgene expression levels in pyramided lines are additive
We measured transgene expression levels of flag leaf samples from the field grown cultivars and lines using non-allele-specific probes in RT qPCR assays. We calculated expression levels relative to Pm3b from line Pm3b as reference, because this line was already used as reference in earlier studies and has an intermediate level of transgene expression (Brunner et al. 2011, 2012). In both years, cultivars Asosan, Chul, and Kolibri carrying the endogenous Pm3 alleles showed very low expression levels (2015: 0.02, 0.01, and 0.03, respectively; 2016: 0.04, 0.01, and 0.04, respectively) compared to line Pm3b (Fig. 5). This is in agreement with the previous results (Brunner et al. 2011, 2012). In both years, parental lines Pm3a and Pm3d showed slightly lower transgene expression levels than line Pm3b with relative scores of 0.38 (Pm3a) and 0.72 (Pm3d) in 2015 and 0.57 (Pm3a) and 0.59 (Pm3d) in 2016 (Fig. 5). In both years, parental line Pm3f showed high transgene expression levels with relative scores of 1.65 in 2015 and 1.98 in 2016 (Fig. 5). All four pyramided lines showed additive total transgene expression levels, i.e., transgene expression levels were the sum of the transgene expression levels of the parental lines (Fig. 5). No suppression among Pm3 alleles at the transcriptional level was observable in any of the pyramided lines (Fig. 5). We then designed allele-specific RT qPCR probes to determine the contribution to transgene expression levels from each of the two Pm3 alleles in the pyramided lines. Overall, the transgene expression levels using the allele-specific probes (Fig. 6) were similar to the data which we obtained using the non-allele-specific probes (Fig. 5). The allele-specific probes revealed that the transgene expression levels of the individual alleles in the pyramided lines were similar to the transgene expression levels of the individual alleles in the parental lines (Fig. 6), resulting in additive transgene expression levels.
Pm3 expression levels determined by RT qPCR using conserved probes (i.e., no distinction between Pm3 alleles) of the field grown pyramided lines (dark green), parental lines (light green), and cultivars Asosan, Chul, and Kolibri (grey) carrying the endogenous alleles Pm3a, Pm3b, and Pm3d, respectively. Expression levels were calculated relative to expression level of Pm3b from field grown line Pm3b. Mean and standard deviation of five independent replicates (pooled flag leaf samples from three plants per plot, from five microplots) are indicated of lines and cultivars grown in 2015 (a) and 2016 (b) (color figure online)
Pm3 expression levels determined by RT qPCR using allele-specific probes. Expression levels are calculated relative to expression level of Pm3b from field grown line Pm3b. Mean and standard deviation of five independent replicates (pooled flag leaf samples from three plants per plot, from five plots) are indicated of lines grown in field season 2015 (a–d) and 2016 (e–h)
Discussion
Combination of allele specificities partially explains improved powdery mildew resistance in pyramided lines
All four pyramided lines were more powdery mildew resistant than their corresponding parental lines in both field seasons (Fig. 1). There are two explanations for resistance, one is related to characteristics of the used plant genotypes and the other is related to characteristics of the pathogen population. In 2015 and 2016, the Bgt population present at the site of the field trial consisted at least partially of strains virulent on all four alleles (Pm3a, b, d, and f) present in the pyramided lines (Fig. 1). Possibly, Bgt strains virulent on Pm3a and Pm3d were more abundant at the site of the field trial than strains virulent on Pm3b and Pm3f. This would partially explain the higher powdery mildew infections on parental lines Pm3a and Pm3d as well as on the pyramided line Pm3a × Pm3d. Earlier studies on virulence frequency of Swiss powdery mildew races to Pm3 alleles present at the field site at Zurich-Reckenholz in 2007 showed that Bgt strains virulent on Pm3f (73% virulence frequency) were more frequent than Bgt strains virulent on Pm3a (6%), Pm3b (4%), and Pm3d (10%) (Brunner et al. 2012). Our results suggest a shift in the Bgt population since 2007, resulting in a much lower frequency of Bgt strains virulent on Pm3f. However, the high powdery mildew resistance of line Pm3f is probably due to other factors than the pathogen population composition, because cultivar Michigan Amber/8*CC which contains the endogenous Pm3f allele, showed powdery mildew infection.
Much has been reported about additive (Hu et al. 2012; Kim et al. 2012; Liu et al. 2000; Xiao et al. 2017) or negative (Chen et al. 2013; Knott 2000; Liu et al. 2013; McIntosh et al. 2011) gene action in various plant species with resistance gene combinations. In these studies, resistance gene combination was achieved by classical crossbreeding and marker-assisted selection or in the case of Chen et al. (2013) by generation of interspecific crosses between tetraploid wheat and Ae. tauschii. Additive or negative gene action has also been reported from resistance gene combinations achieved by genetic engineering resulting in trans- or cisgenic plants. Additive gene action was reported for transgenic flax with pyramided rust resistance alleles (Chen et al. 2008) and for transgenic potatoes with pyramided late blight-resistance genes (Zhu et al. 2012; Jo et al. 2014), and suppression was reported from transgenic wheat carrying Pm3 and Pm8 (Hurni et al. 2014). However, no field trials were performed with the transgenic flax lines or the transgenic wheat lines carrying Pm3 and Pm8. Tundo et al. (2016) pyramided various genes encoding different inhibitors to generate transgenic wheat resistant to Fusarium graminearum. Some inhibitor combinations improved the resistance spectrum (Tundo et al. 2016). In the case of potatoes with improved resistance spectra against Phytophthora infestans, both classical crossbreeding with marker-assisted selection and genetic engineering have been used and field trials were conducted. The two resistance genes RPi-mcd1 and RPi-ber were combined by crossbreeding and field trials revealed an additive effect of the two genes (Tan et al. 2010). Furthermore, potato differential lines MaR8 and MaR9 show broad spectrum late blight resistance due to multiple pyramided R genes (Kim et al. 2012). Zhu et al. (2012) transformed potato cultivar Desiree simultaneously with three broad spectrum potato R genes which resulted in a resistance spectrum which was the sum of the spectra from the three individual R genes (Zhu et al. 2012). Jo et al. (2014) then developed a marker-free transformation procedure to obtain cisgenic potato lines expressing several pyramided R genes (Jo et al. 2014). The cisgenic potato lines showed improved resistance in field trials (Haverkort et al. 2016; Haesaert et al. 2015). Thus, in potato, the pyramiding of R genes is a successful approach to broaden the resistance spectrum, as seems to be the case for our Pm3 allele-pyramided wheat lines.
Resistance suppression among pyramided Pm3 alleles observed in laboratory experiments was not confirmed in the field
Stirnweis et al. (2014) tested powdery mildew resistance of the four pyramided lines used in this study in infection tests in the laboratory on leaf segments of 10-day-old seedlings. When Bgt isolates 97011 and 98229 which were used for infection, a resistance suppression was observed, namely Pm3 variants Pm3b and Pm3c suppressed the function of both Pm3 variants Pm3a and Pm3f. Surprisingly, we did not observe functional suppression of Pm3 variants Pm3a or Pm3f by variant Pm3b in the field grown plants; on the contrary, pyramided lines Pm3a × Pm3b and Pm3b × Pm3f were completely powdery mildew resistant in both years with AUDPC scores of 0 (Fig. 1). Stirnweis et al. (2014) observed the suppression phenotype of the allele-pyramided lines only when using Bgt isolates 97011 (avirulent on Pm3a/d/f, virulent on Pm3b/c) and 98229 (avirulent on Pm3a/d/f, virulent on Pm3b/c) but not when using isolates Bgt 07201 (avirulent on Pm3b, virulent on Pm3a/f) and 07230 (avirulent on Pm3b/c, virulent on Pm3a/f). The infection tests were performed on 10-day-old wheat seedlings. The disparate results from the seedling tests and the field trials cannot be attributed to the age difference of the plants, because it is known that Pm3 already is effective at the 10-day-old seedling stage (Brunner et al. 2010, 2011, 2012; Stirnweis et al. 2014). Stirnweis et al. (2014) further investigated the suppression mechanism and concluded it occurred at the post-translational level, because RNA and protein levels of Pm3 alleles/variants were unaffected by the presence of suppressing Pm3 alleles/variants. In transiently transformed N. benthamiana leaves expressing Pm3b and autoactive Pm3f-D501V, Stirnweis et al. (2014) observed suppression of the hypersensitive response, which suggests that in the presence of activated Pm3f, the suppression is independent of the presence of Bgt isolates. Our result of the absence of observable allele suppression in the same pyramided lines when grown in field conditions adds to the puzzle of the underlying mechanism of Pm3 activation and function. Further molecular studies of laboratory and field grown plants infected with various Bgt isolates are necessary to elucidate the observed resistance or suppression phenotypes. At this point, we only can speculate on possible factors explaining the disparate results obtained in laboratory and field trials. One factor, besides the Bgt population composition, could be a difference in immune system activity of laboratory and field grown plants; for instance, an avirulent Bgt isolate landing on a field grown plant could prime the plant for improved immunity and subsequent Bgt strains virulent under laboratory conditions could remain avirulent under field conditions. In our field trial, besides the naturally occurring Bgt population, Bgt isolate 96224, which is avirulent on all field tested Pm3 alleles, was present in the field because of the Bgt 96224 infected spreader rows.
Positive correlation between total transgene expression level and powdery mildew resistance in transgenic wheat lines of cultivar Bobwhite
In the field grown parental and pyramided lines, all in the background of cultivar Bobwhite, we observed a positive correlation between total transgene expression level and level of powdery mildew resistance. The three lines Pm3b × Pm3f, Pm3a × Pm3b, and Pm3f had the highest total transgene expression levels in both years and where completely powdery mildew resistant in both years, whereas lines Pm3a and Pm3d were quite susceptible in both years and showed the lowest transgene expression levels in both years (Figs. 1, 5). Only few studies so far investigate the correlation between field resistance and transgene expression levels. Han et al. (2012) reported a positive correlation of the level of transgene expression (cDNA of bovine lactoferrin) and resistance of wheat to F. graminearum (Han et al. 2012). Krattinger et al. (2016) reported a positive correlation between level of transgene expression of resistance gene Lr34 in rice and level of leaf tip necrosis, but transgene expression level was not positively correlated to disease resistance (Krattinger et al. 2016). Both these studies did not conduct field trials with the transgenic plants. High levels of transgene expression in plants can lead to gene silencing (Pickford and Cogoni 2003). However, we did not observe gene silencing in any of our field tested lines. Kim et al. (2012) reported a background dependence of resistance gene action in potato. In our study, allele Pm3d performed better in the endogenous background of cultivar Kolibri than in our Bobwhite Pm3d overexpressing line, even though Pm3d was much higher expressed in Bobwhite line Pm3d than in Kolibri (Fig. 5). Cultivar Kolibri has been used agronomically in Switzerland since 1975. In contrast to cultivar Bobwhite, Kolibri is locally adapted and thus is likely to carry additional, unknown powdery mildew resistance factors (e.g., minor, quantitative resistance genes), which are absent in cultivar Bobwhite. Thus, even though we observed a positive correlation between transgene expression level and powdery mildew resistance in transformed cultivar Bobwhite, Pm3 expression levels alone do not explain strength of powdery mildew resistance.
Pm3 allele pyramiding is a successful approach for enhancing powdery mildew field resistance
Overactive immunity in plants can have negative effects on plant development (Belkhadir et al. 2014; Lozano-Durán and Zipfel 2015). We did not observe a correlation of total transgene expression level and abnormal plant development. On the contrary, pyramided line Pm3b × Pm3f, which had a higher total transgene expression level than parental line Pm3f, showed yield, flowering date, and seed set rate more similar to non-transformed Bobwhite than to parental line Pm3f, which had an abnormal plant development phenotype (Figs. 2, 4, 5, 6). The observed severely delayed flowering and low seed set rate of line Pm3f, but not of sister line Pm3f, is in agreement with Brunner et al. (2012). In the case of pyramided line Pm3b × Pm3f, crossbreeding of “abnormal” line Pm3f with line Pm3b to a large extent restored normal plant development. The other three pyramided lines did not show any negative effects of allele pyramiding on plant development neither. Pm3 allele pyramiding did not have a negative effect on the plants and was a successful approach in enhancing field resistance to powdery mildew. Furthermore, it was a more successful approach than using line mixtures (Fig. 3b). Brunner et al. (2012) showed improved powdery mildew resistance of the multilines compared to the lines in pure stands. In our study, we showed further improvement of resistance by allele pyramiding, which, in the two pyramided lines Pm3a × Pm3b and Pm3b × Pm3f, resulted in plants completely free of powdery mildew infection. We attribute the improved resistance to the two effects of enhanced total transgene expression levels and allele-specificity combinations. The transgenic lines used in this study could be further improved with characteristics of interest by classical crossbreeding.
Author contribution statement
SB, SH, BK, and TK designed and carried out the field trial; GH designed and carried out the RT qPCR experiments; TK, SB, and BK wrote the manuscript.
References
Belkhadir Y, Yang L, Hetzel J, Dangl JL, Chory J (2014) The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors. Trends Biochem Sci 39:447–456
Bhullar NK, Street K, Mackay M, Yahiaoui N, Keller B (2009) Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc Natl Acad Sci USA 106:9519–9524
Bhullar NK, Zhang Z, Wicker T, Keller B (2010) Wheat gene bank accessions as a source of new alleles of the powdery mildew resistance gene Pm3: a large scale allele mining project. BMC Plant Biol 10:88
Bourras S, McNally KE, Ben-David R, Parlange F, Roffler S, Praz CR, Oberhaensli S, Menardo F, Stirnweis D, Frenkel Z, Schaefer LK, Fluckiger S, Treier G, Herren G, Korol AB, Wicker T, Keller B (2015) Multiple avirulence loci and allele-specific effector recognition control the Pm3 Race-specific resistance of wheat to powdery mildew. Plant Cell 27:2991–3012
Bourras S, McNally KE, Müller MC, Wicker T, Keller B (2016) Avirulence genes in cereal powdery mildews: the gene-for-gene hypothesis 2.0. Front Plant Sci 7:241
Brunner S, Hurni S, Streckeisen P, Mayr G, Albrecht M, Yahiaoui N, Keller B (2010) Intragenic allele pyramiding combines different specificities of wheat Pm3 resistance alleles. Plant J 64:433–445
Brunner S, Hurni S, Herren G, Kalinina O, von Burg S, Zeller SL, Schmid B, Winzeler M, Keller B (2011) Transgenic Pm3b wheat lines show resistance to powdery mildew in the field. Plant Biotechnol J 9:897–910
Brunner S, Stirnweis D, Diaz Quijano C, Buesing G, Herren G, Parlange F, Barret P, Tassy C, Sautter C, Winzeler M, Keller B (2012) Transgenic Pm3 multilines of wheat show increased powdery mildew resistance in the field. Plant Biotechnol J 10:398–409
Burdon JJ, Barrett LG, Rebetzke G, Thrall PH (2014) Guiding deployment of resistance in cereals using evolutionary principles. Evol Appl 7:609–624
Chen Y, Singh S, Rashid K, Dribnenki P, Green A (2008) Pyramiding of alleles with different rust resistance specificities in Linum usitatissimum L. Mol Breed 21:419–430
Chen W, Liu T, Gao L (2013) Suppression of stripe rust and leaf rust resistances in interspecific crosses of wheat. Euphytica 192:339–346
Delmotte F, Bourguet D, Franck P, Guillemaud T, Reboud X, Vacher C, Walker A-S (2016) Combining selective pressures to enhance the durability of disease resistance genes. Front Plant Sci 7:1916
Haesaert G, Vossen JH, Custers R, De Loose M, Haverkort A, Heremans B, Hutten R, Kessel G, Landschoot S, Van Droogenbroeck B, Visser RGF, Gheysen G (2015) Transformation of the potato variety Desiree with single or multiple resistance genes increases resistance to late blight under field conditions. Crop Prot 77:163–175
Han J, Lakshman DK, Galvez LC, Mitra S, Baenziger PS, Mitra A (2012) Transgenic expression of lactoferrin imparts enhanced resistance to head blight of wheat caused by Fusarium graminearum. BMC Plant Biol 12:33
Haverkort AJ, Boonekamp PM, Hutten R, Jacobsen E, Lotz LAP, Kessel GJT, Vossen JH, Visser RGF (2016) Durable late blight resistance in potato through dynamic varieties obtained by cisgenesis: scientific and societal advances in the DuRPh Project. Potato Res 59:35–66
Hillel J, Schaap T, Haberfeld A, Jeffreys AJ, Plotzky Y, Cahaner A, Lavi U (1990) DNA fingerprints applied to gene introgression in breeding programs. Genetics 124:783–789
Hu J, Li X, Wu C, Yang C, Hua H, Gao G, Xiao J, He Y (2012) Pyramiding and evaluation of the brown planthopper resistance genes Bph14 and Bph15 in hybrid rice. Mol Breed 29:61–69
Hurni S, Brunner S, Stirnweis D, Herren G, Peditto D, McIntosh RA, Keller B (2014) The powdery mildew resistance gene Pm8 derived from rye is suppressed by its wheat ortholog Pm3. Plant J 79:904–913
Hurni S, Scheuermann D, Krattinger SG, Kessel B, Wicker T, Herren G, Fitze MN, Breen J, Presterl T, Ouzunova M, Keller B (2015) The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc Natl Acad Sci 112:8780–8785
Jo K-R, Kim C-J, Kim S-J, Kim T-Y, Bergervoet M, Jongsma M, Visser RGF, Jacobsen E, Vossen JH (2014) Development of late blight resistant potatoes by cisgene stacking. BMC Biotechnol 14:50
Kim H-J, Lee H-R, Jo K-R, Mortazavian SMM, Huigen DJ, Evenhuis B, Kessel G, Visser RGF, Jacobsen E, Vossen JH (2012) Broad spectrum late blight resistance in potato differential set plants MaR8 and MaR9 is conferred by multiple stacked R genes. Theor Appl Genet 124:923–935
Knott DR (2000) Inheritance of resistance to stem rust in Medea durum wheat and the role of suppressors. Crop Sci 40:98–102
Krattinger SG, Sucher J, Selter LL, Chauhan H, Zhou B, Tang M, Upadhyaya NM, Mieulet D, Guiderdoni E, Weidenbach D, Schaffrath U, Lagudah ES, Keller B (2016) The wheat durable, multipathogen resistance gene Lr34 confers partial blast resistance in rice. Plant Biotechnol J 14:1261–1268
Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BP, Staskawicz B, Jones JD, Zipfel C (2010) Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol 28:365–369
Liu J, Liu D, Tao W, Li W, Wang S, Chen P, Cheng S, Gao D (2000) Molecular marker-facilitated pyramiding of different genes for powdery mildew resistance in wheat. Plant Breed 119:21–24
Liu W, Danilova TV, Rouse MN, Bowden RL, Friebe B, Gill BS, Pumphrey MO (2013) Development and characterization of a compensating wheat-Thinopyrum intermedium Robertsonian translocation with Sr44 resistance to stem rust (Ug99). Theor Appl Genet 126:1167–1177
Liu Q, Hallerman E, Peng Y, Li Y (2016) Development of Bt rice and Bt maize in China and their efficacy in target pest control. Int J Mol Sci 17:1561
Lozano-Durán R, Zipfel C (2015) Trade-off between growth and immunity: role of brassinosteroids. Trends Plant Sci 20:12–19
McDonald BA, Linde C (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 40:349–379
McIntosh RA, Zhang P, Cowger C, Parks R, Lagudah ES, Hoxha S (2011) Rye-derived powdery mildew resistance gene Pm8 in wheat is suppressed by the Pm3 locus. Theor Appl Genet 123:359–367
Mondal S, Rutkoski JE, Velu G, Singh PK, Crespo-Herrera LA, Guzmán C, Bhavani S, Lan C, He X, Singh RP (2016) Harnessing diversity in wheat to enhance grain yield, climate resilience, disease and insect pest resistance and nutrition through conventional and modern breeding approaches. Front Plant Sci 7:991
Narusaka M, Kubo Y, Hatakeyama K, Imamura J, Ezura H, Nanasato Y, Tabei Y, Takano Y, Shirasu K, Narusaka Y (2013) Interfamily transfer of dual NB-LRR genes confers resistance to multiple pathogens. PLoS One 8:e55954
Parlange F, Roffler S, Menardo F, Ben-David R, Bourras S, McNally KE, Oberhaensli S, Stirnweis D, Buchmann G, Wicker T, Keller B (2015) Genetic and molecular characterization of a locus involved in avirulence of Blumeria graminis f. sp. tritici on wheat Pm3 resistance alleles. Fungal Genet Biol 82:181–192
Pickford AS, Cogoni C (2003) RNA-mediated gene silencing. Cell Mol Life Sci 60:871–882
Rodriguez-Moreno L, Song Y, Thomma BP (2017) Transfer and engineering of immune receptors to improve recognition capacities in crops. Curr Opin Plant Biol 38:42–49
Romeis J, Meissle M, Brunner S, Tschamper D, Winzeler M (2013) Plant biotechnology: research behind fences. Trends Biotechnol 31:222–224
Srichumpa P, Brunner S, Keller B, Yahiaoui N (2005) Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol 139:885–895
Stirnweis D, Milani SD, Brunner S, Herren G, Buchmann G, Peditto D, Jordan T, Keller B (2014) Suppression among alleles encoding nucleotide-binding-leucine-rich repeat resistance proteins interferes with resistance in F1 hybrid and allele-pyramided wheat plants. Plant J 79:893–903
Tan MYA, Hutten RCB, Visser RGF, van Eck HJ (2010) The effect of pyramiding Phytophthora infestans resistance genes R Pi-mcd1 and R Pi-ber in potato. Theor Appl Genet 121:117–125
Tanksley SD, Nelson JC (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92:191–203
Tundo S, Kalunke R, Janni M, Volpi C, Lionetti V, Bellincampi D, Favaron F, D’Ovidio R (2016) Pyramiding PvPGIP2 and TAXI-III but not PvPGIP2 and PMEI enhances resistance against Fusarium graminearum. Mol Plant Microbe Interact 29:629–639
Visscher PM, Haley CS, Thompson R (1996) Marker-assisted introgression in backcross breeding programs. Genetics 144:1923–1932
Xiao N, Wu Y, Pan C, Yu L, Chen Y, Liu G, Li Y, Zhang X, Wang Z, Dai Z, Liang C, Li A (2017) Improving of rice blast resistances in japonica by pyramiding major R genes. Front Plant Sci 7:1–10
Yahiaoui N, Srichumpa P, Dudler R, Keller B (2004) Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 37:528–538
Yahiaoui N, Brunner S, Keller B (2006) Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J 47:85–98
Zhan J, Thrall PH, Papaix J, Xie L, Burdon JJ (2015) Playing on a Pathogen’ s weakness: using evolution to guide sustainable plant disease control strategies. Annu Rev Phytopathol 53:19–43
Zhu S, Li Y, Vossen JH, Visser RGF, Jacobsen E (2012) Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Res 21:89–99
Acknowledgements
We would like to thank Mario Waldburger, Agroscope Zurich-Reckenholz for photographs, Linda Lüthi, Christine Meran, Anna Landwehr, Valentino De Conno, Helen Zbinden, and Bea Senger as well as the staff from Agroscope in Zurich-Reckenholz for their help in the field. We thank Anne Roulin and Javier Sanchez-Martin for their help with statistics. Funding was provided by a Grant from the Swiss National Science Foundation 310030_163260 and by Forschungskredit Grant No. FK-15-098 from the University of Zurich to TK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Xiaoquan Qi.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Koller, T., Brunner, S., Herren, G. et al. Pyramiding of transgenic Pm3 alleles in wheat results in improved powdery mildew resistance in the field. Theor Appl Genet 131, 861–871 (2018). https://doi.org/10.1007/s00122-017-3043-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-3043-9