Abstract
Circular RNAs (circRNAs) are well-known to exert significant roles in regulating the pathological processes, including human carcinogenesis. Currently, less is known about their exact roles in head and neck squamous cell carcinoma (HNSCC). Herein, we aimed to investigate and validate the role of a novel circRNA, circMAT2B, as well as its potential molecular mechanism in HNSCC progression. A cohort of 41 paired of HNSCC tumor tissues and adjacent normal tissues from HNSCC patients were collected. Further, we characterized circMAT2B expression patterns in HNSCC tissues and cell lines, as well as exploring its association with the prognosis of HNSCC patients. Biological functions on cell proliferation, apoptosis, migration, and invasion were assessed using Cell Counting Kit-8, EdU incorporation, TUNEL, wound healing, and transwell assays. Glutaminolysis was evaluated by measuring glutamine, glutamate, and α-ketoglutarate (α-KG) levels. The regulatory network of circMAT2B/miR-491-5p/ASCT2 axis was verified by RNA immunoprecipitation and luciferase reporter assays. Western blot was conducted to detect the level of ASCT2 and GLS1. Remarkably overexpressed circMAT2B was observed in HNSCC tissues and cell lines, of which high abundance was positively correlated with patients’ poor prognosis. Silencing of circMAT2B inhibited cell proliferation, migration, and invasion, as well as glutaminolysis. miR-491-5p, interacted with ASCT2, was identified to be a downstream target of circMAT2B, thereby involving in circMAT2B-mediated biological effects. In summary, we draw a conclusion that circMAT2B could modulate the processes of cell proliferation, migration, invasion, and glutaminolysis of HNSCC cells partly via the miR-491-5p/ASCT2 axis by a molecular mechanism of competing endogenous RNA (ceRNA), implying an underlying circRNA-targeted therapy for HNSCC treatment.








Similar content being viewed by others
Data availability
Enquiries about data availability should be directed to the authors.
References
Hsu MT, Coca-Prados M (1979) Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280:339–340
Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA 73:3852–3856
Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H (1986) The hepatitis delta (delta) virus possesses a circular RNA. Nature 323:558–560
Cocquerelle C, Mascrez B, Hetuin D, Bailleul B (1993) Mis-splicing yields circular RNA molecules. FASEB J 7:155–160
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338
Liu L, Wang J, Khanabdali R, Kalionis B, Tai X, Xia S (2017) Circular RNAs: isolation, characterization and their potential role in diseases. RNA Biol 14:1715–1721
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20:675–691
Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP (2016) Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 165:289–302
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S (2015) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25:981–984
Zhu Z, Liu Y, Wu D, Wang H (2021) Association between mitochondrial DNA Copy number and head and neck squamous cell carcinoma: a systematic review and dose-response meta-analysis. Med Sci Monit 27:e928327
Vergeer MR, Doornaert PA, Rietveld DH, Leemans CR, Slotman BJ, Langendijk JA (2009) Intensity-modulated radiotherapy reduces radiation-induced morbidity and improves health-related quality of life: results of a nonrandomized prospective study using a standardized follow-up program. Int J Radiat Oncol Biol Phys 74:1–8
Leemans CR, Braakhuis BJ, Brakenhoff RH (2011) The molecular biology of head and neck cancer. Nat Rev Cancer 11:9–22
Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X (2019) Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology 70:1298–1316
Liu J, Liu H, Zeng Q, Xu P, Liu M, Yang N (2020) Circular RNA circ-MAT2B facilitates glycolysis and growth of gastric cancer through regulating the miR-515-5p/HIF-1alpha axis. Cancer Cell Int 20:171
Mates JM, Campos-Sandoval JA, Santos-Jimenez JL, Marquez J (2019) Dysregulation of glutaminase and glutamine synthetase in cancer. Cancer Lett 467:29–39
Cluntun AA, Lukey MJ, Cerione RA, Locasale JW (2017) Glutamine metabolism in cancer: understanding the heterogeneity. Trends Cancer 3:169–180
Hensley CT, Wasti AT, DeBerardinis RJ (2013) Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest 123:3678–3684
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, Shu Y (2019) CircRNAs in cancer metabolism: a review. J Hematol Oncol 12:90
Akins NS, Nielson TC, Le HV (2018) Inhibition of Glycolysis and glutaminolysis: an emerging drug discovery approach to combat cancer. Curr Top Med Chem 18:494–504
Yang L, Venneti S, Nagrath D (2017) Glutaminolysis: a hallmark of cancer metabolism. Annu Rev Biomed Eng 19:163–194
Kamarajan P, Rajendiran TM, Kinchen J, Bermudez M, Danciu T, Kapila YL (2017) Head and neck squamous cell carcinoma metabolism draws on glutaminolysis, and stemness is specifically regulated by glutaminolysis via aldehyde dehydrogenase. J Proteome Res 16:1315–1326
Guo Y, Yang J, Huang Q, Hsueh C, Zheng J, Wu C, Chen H, Zhou L (2019) Circular RNAs and their roles in head and neck cancers. Mol Cancer 18:44
Dai F, Dai L, Zheng X, Guo Y, Zhang Y, Niu M, Lu Y, Li H, Hou R, Zhang Y, Wen S, Hu W, An C, Wu Y, Gao W (2020) Non-coding RNAs in drug resistance of head and neck cancers: a review. Biomed Pharmacother 127:110231
Wang H, Feng L, Cheng D, Zheng Y, Xie Y, Fu B (2021) Circular RNA MAT2B promotes migration, invasion and epithelial-mesenchymal transition of non-small cell lung cancer cells by sponging miR-431. Cell Cycle 20:1617–1627
Zhao JP, Chen LL (2020) Circular RNA MAT2B induces colorectal cancer proliferation via sponging mir-610, resulting in an increased E2F1 expression. Cancer Manag Res 12:7107–7116
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, Wei F, Guo C, Wu X, Li X, Li Y, Li G, Zeng Z, Xiong W (2018) Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 17:79
Liu F, Zhang H, Xie F, Tao D, Xiao X, Huang C, Wang M, Gu C, Zhang X, Jiang G (2020) Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene 39:1696–1709
Guo J, Luo C, Yang Y, Dong J, Guo Z, Yang J, Lian H, Ye C, Liu M (2021) MiR-491-5p, as a tumor suppressor, prevents migration and invasion of breast cancer by targeting ZNF-703 to regulate AKT/mTOR pathway. Cancer Manag Res 13:403–413
Lu L, Cai M, Peng M, Wang F, Zhai X (2019) miR-491-5p functions as a tumor suppressor by targeting IGF2 in colorectal cancer. Cancer Manag Res 11:1805–1816
Chen T, Li Y, Cao W, Liu Y (2018) miR-491-5p inhibits osteosarcoma cell proliferation by targeting PKM2. Oncol Lett 16:6472–6478
Wu F, Ji A, Zhang Z, Li J, Li P (2021) miR-491-5p inhibits the proliferation and migration of A549 cells by FOXP4. Exp Ther Med 21:622
Yu T, Wang LN, Li W, Zuo QF, Li MM, Zou QM, Xiao B (2018) Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Sci 109:1393–1403
Kang W, Zhang J, Huang T, Zhou Y, Wong CC, Chan RCK, Dong Y, Wu F, Zhang B, Wu WKK, Chan MWY, Cheng ASL, Yu J, Wong N, Lo KW, To KF (2021) NOTCH3, a crucial target of miR-491-5p/miR-875-5p, promotes gastric carcinogenesis by upregulating PHLDB2 expression and activating Akt pathway. Oncogene 40:1578–1594
Reddy KB (2015) MicroRNA (miRNA) in cancer. Cancer Cell Int 15:38
Kanai Y, Hediger MA (2004) The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pflugers Arch 447:469–479
van Geldermalsen M, Wang Q, Nagarajah R, Marshall AD, Thoeng A, Gao D, Ritchie W, Feng Y, Bailey CG, Deng N, Harvey K, Beith JM, Selinger CI, O’Toole SA, Rasko JE, Holst J (2016) ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene 35:3201–3208
Schulte ML, Fu A, Zhao P, Li J, Geng L, Smith ST, Kondo J, Coffey RJ, Johnson MO, Rathmell JC, Sharick JT, Skala MC, Smith JA, Berlin J, Washington MK, Nickels ML, Manning HC (2018) Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat Med 24:194–202
Zhang Z, Liu R, Shuai Y, Huang Y, Jin R, Wang X, Luo J (2020) ASCT2 (SLC1A5)-dependent glutamine uptake is involved in the progression of head and neck squamous cell carcinoma. Br J Cancer 122:82–93
Funding
This study was supported by Natural Science Foundation of China (No. 82073006, No. 81902757, No. 82002757).
Author information
Authors and Affiliations
Contributions
J-TL: Conceptualization; Writing-original draft; Methodology; Formal analysis; Y-fW: Supervision; Validation; YW, R-YL: Data curation; Resources; C-LW: Investigation; ZZ: Funding acquisition; Project administration; Writing-review & editing. All authors have read and approved the final version of this manuscript to be published.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval
Patients enrolled in this study signed their informed consent for participation and our work was approved by the Ethics Committee of Tianjin Cancer Hospital in accordance to the Declaration of Helsinki.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11010_2022_4565_MOESM1_ESM.tif
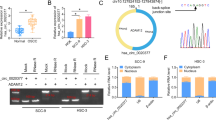
Supplementary file1 (TIF 558 kb)— Identification of circMAT2B originated from MAT2B. (A) Schematic diagram illustrated the formation of circMAT2B originated from MAT2B pre-mRNA (exon 3, 4, 5, 6). (B) RNA stability assay was performed to detect the expression level of circMAT2B or linear mRNA transcripts in CAL27 cells under the exposure of transcription inhibitor (actinomycin D). (C) qRT-PCR analysis was performed to determine the expression of circMAT2B and MAT2B mRNA in CAL27 cells administered with RNase R or Mock control. Data were representative images or were expressed as the mean ± SD of n = 3 experiments. *, P < 0.05, **, P < 0.01, ***, P < 0.001
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Luo, JT., Wang, Yf., Wang, Y. et al. CircMAT2B facilitates the progression of head and neck squamous cell carcinoma via sponging miR-491-5p to trigger ASCT2-mediated glutaminolysis. Mol Cell Biochem 478, 1067–1081 (2023). https://doi.org/10.1007/s11010-022-04565-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-022-04565-3