Abstract
Altered energy metabolism is a hallmark of tumors aiming at supplying necessary nutrients for tumorigenesis and development. These redirected metabolic pathways associated with carbohydrate, lipid and amino acid are orchestrated not only by carcinogenic proteins but by non-coding RNAs. Among them, circular RNA (circRNA), as a kind of novel identified non-coding RNAs, has become the focus of attention. Through binding with corresponding microRNAs or directly contacting proteins, circRNA plays a primarily important role in regulating cellular metabolism. Herein, we analyze the emerging findings and select circRNAs contributing to mutant glycolysis, lipogenesis and lipolysis, glutam inolysis, and oxidative respiration to deepen the understanding about the cancer metabolic regulatory network. In addition, we also discuss the possibility of circRNAs exerting their functions via exosomes and cancer stem cells. Owing to their unique structures and wide impacts, circRNAs may help reap huge fruits in developing clinical treatments targeting cancer metabolism.
Similar content being viewed by others
Background
Reprogramming of energy metabolism is a hallmark of tumors caused by genome instability [1]. Deregulated metabolism, which is widespread in tumor progression, provides an essential source for growth and division of cancer cells. Compared with normal adult tissues, carbonhydrate, lipid, and amino acid metabolisms may undergo dramatic transformational changes in tumors (Fig. 1). The recognized cancer-associated metabolic changes include deregulated uptake of glucose and amino acids, use of opportunistic modes of nutrient acquisition, use of glycolysis/tricarboxylic acid cycle (TCA), cycle intermediates for biosynthesis and nicotinamide adenine dinucleotide phosphate (NADPH) production, and increased demand for nitrogen [2]. To develop effective therapeutic strategies for cancers, it is of pivotal importance to investigate the mechanism underlying cancer abnormal metabolism.
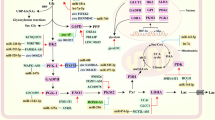
Schematic representation of altered metabolic pathways and associated circRNAs in cancer. The characteristic features include two aspects. First, it illustrates deregulated uptake of glucose and amino acids, overactive fatty acid synthesis and oxidation, and NADPH overproduction. Second, it shows how circRNAs interact with these metabolic pathways by enzymes and TFs. HIF1-α and c-myc can adjust the Warburg effect-relevant enzymes and transporters. The majority of circRNAs can antagonize miRNAs to indirectly produce effects. In addition, circ-Amotl1 and circ_FBXW7 can directly affect TFs. The circRNAs in the dark blue rounded rectangle are considered to function in cancer cells, while the circRNAs in the light blue rounded rectangle are just proved to function in normal cells. miRNAs are in the green rectangle; the components affecting metabolism are in faint yellow rectangle. The positive relationships are shown by arrows, and the negative relationships are shown by short dashes. ACC, acetyl-COA carboxylase; ACSL, acyl-CoA synthetases; ASCT2, neutral amino acid transporter2; BC, breast cancer; CPT1, carnitine palmitoyltransferase 1; CRC, colorectal cancer; FAO, fatty acids β-oxidation; FASN, fatty acid synthase; FAT, fatty acid translocase; GLS, glutaminase; GLUD1, glutamate dehydrogenase; GLUT, glutamate dehydrogenase; HCC, hepatocellular carcinoma; HK, hexokinase; LDHA, lactate dehydrogenase A; LUAD, lung adenocarcinoma; MCT. monocarboxylate transporter; PCa, prostate cancer; PDH, pyruvate dehydrogenase; PDK, pyruvate dehydrogenase kinase; PFK, 6-phosphfructa-1-kinase; PKM2, Pyruvate kinase isozymes M2; PPP, pentose phosphate pathway; α-KG, α-ketoglutarate. (Red lines, catabolic pathways; orange lines, glutamine pathways; green lines, lipid pathways)
These redirections of energy metabolism are orchestrated by both proteins and non-coding RNAs (ncRNAs). As an emerging category of regulatory molecules, ncRNA has been verified to control cancer metabolism [3]. For instance, long non-coding RNA PCGEM1 can promote glucose uptake for aerobic glycolysis in prostate cancer (PCa) cells [4]. Similarly, circular RNA (circRNA) as another type of ncRNA may also get entangled with cancer metabolism.
CircRNA is a kind of ncRNA whose structure comprises covalently closed loops without either poly-adenylated tails in 3′ ends or the cap structure at 5′ ends. Due to this extraordinary construction, circRNAs can avoid exonuclease degradation and have much longer half-life than their parental mRNAs (48 h vs. 10 h) [5]. According to this conservative property, many studies have focused their attention on the potential role of circRNAs as a promising disease biomarker. Besides, accumulating investigations have found the regulatory functions of circRNAs. They can directly regulate transcription by interacting with mRNAs or long non-coding RNAs (lncRNAs), sponging mRNAs, or RNA binding proteins (RBPs) [6, 7]. Some circRNAs can even be translated into proteins [8]. It is rational to hypothesize that circRNAs may regulate cancer metabolism through sponging miRNAs or other targets. Hence, in this article, we collect putative circRNAs affecting metabolism and divide them into three groups associated with carbohydrate, lipid, and amino acid respectively. The aim of this review is to gain insights into the relationship between circRNAs and cancer metabolism and provide a better theoretical basis for the clinical diagnosis and treatment of cancers.
Main text
circRNAs in glucose metabolism
The modification of glucose metabolism is part and parcel of the most distinct differentiation in cancer. Normally, cells consume glucose to generate ATP through oxidative phosphorylation (OXPHOS) under the aerobic condition and channel into glycolysis only under the anoxic condition. However, cancer cells prefer glycolysis even in the normoxic environment. This phenomenon is known as “the Warburg effect” as Otto Warburg reported this phenomenon in the 1920s [9]. The Warburg effect is instrumental in malignancy. First, the change from OXPHOS to glycolysis relieves oxidative stress injury caused by mitochondria [10]. Second, excess lactic acid produced by the Warburg effect helps avoid immune surveillance. Besides, it is worth noting that recent profiles have proved that the Warburg effect does not account for the main energy supply due to the inefficiency of glycolysis in ATP synthesis. So it is hypothesized that the Warburg effect consumes a myriad of glucose to either create a hyperacid microenvironment or just starve normal cells in the vicinity [11]. The produced lactate can also stimulate vascularization via HIF1-α [12]. Specifically, the Warburg effect is triggered by abnormal expression of glucose transporters, metabolic enzymes, and oncogenes. Several important signaling pathways also participate in the regulation of this process. Hereinafter, we illustrate the effects of circRNAs on aerobic glycolysis of all these levels (Table 1).
circRNAs play a vital role in glycolysis by regulating transporters and enzymes
As the carbohydrate metabolism is a major energy-providing pathway, a group of metabolic enzymes work together to maintain successful running of the process. Firstly, the glucose transporter (GLUT) takes charge of the import of glucide. To meet the excessive requirements of glucose, GLUTs ranging from GLUT1 to GLUT4 are all overexpressed in tumor cells. Then, three speed-limiting enzymes including hexokinase (HK), 6-phosphfructa-1-kinase (PFK), and pyruvate kinase (PK) work together to transform glucose into pyruvate. Lactate dehydrogenase A (LDHA), which executes the final step of aerobic glycolysis, can catalyze pyruvate into lactate. Finally, the monocarboxylate transporter (MCT) releases lactate into the extracellular matrix. In addition, pyruvate dehydrogenase kinase (PDK) blocks the traditional OXPHOS by countering pyruvate dehydrogenase (PDH) that catalyzes pyruvate into acetyl-COA [29]. PDK1 is a component of the PI3K/AKT pathway to affect cell growth, differentiation, and survival [30].
Some circRNAs can affect these enzymes. Some experimental studies reported that silencing of circHIPK3, which is abundant in pancreatic islets, decreased Slc2a2 expression that encodes GLUT2 [13]. CircHIPK3 could sponge miR-124, which represses the expression of several enzymes and transporters of glycolysis [31, 32]. Another circRNA circ-Amotl1 was reported to be able to physically bind to PDK1 and AKT1 and translocate them into the nucleus in order to antagonize apoptosis [14]. These circRNAs are likely to induce the Warburg effect in tumor tissues.
circRNAs affect glycolysis by regulating transcription factors
Metabolic changes in cancer are viewed as a subsequent step of the molecular transformation process. Distinctive transcription factors (TFs) also affect the Warburg effect. As a member of ncRNAs, circRNA regulates the mutant expression of TFs. So we pay attention to the selected TFs to elucidate the relationships between circRNAs and TFs (Fig. 2).
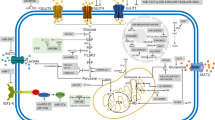
circRNAs and the Warburg effect associated signaling pathways. This figure indicates the effects of circRNAs on the Warburg effect-associated signaling pathways. The PI3K/Akt pathway promotes GLUT1, HK2, and PFK2. The alteration of RAS pathway upregulates GLUT1 and glycolytic enzymes. The STAT3 pathway gives rise to HK2. And the Wnt/β-catenin pathway upregulates the transcription of MCT1 and PDK1. In these four pathways, most circRNAs work as ceRNAs to affect the Warburg effect. The circRNAs in the dark blue rounded rectangle are considered to function in cancer cells, while the circRNAs in the light blue rounded rectangle are just proved to function in normal cells. miRNAs are in the green rectangle; the components of the PI3K/Akt pathway are in the pink circle; the components of the RAS pathway are in the orange circle; the components of the STAT3 pathway are in the green circle; the components of the Wnt/β-catenin pathway are in the yellow circle. The positive relationships are shown by arrows, and the negative relationships are shown by short dashes. BLC, bladder cancer; CRC, colorectal cancer; GC, gastric cancer; OS, osteosarcoma; OSCC, oral squamous cell carcinoma ; PTC, papillary thyroid cancer
HIF-1
There is a tight association between hypoxia and the Warburg effect [33]. Solid tumors encountering hypoxia have to adopt glycolysis instead of oxidative metabolism. In response to the hypoxic condition, HIF-1 contributes to the overexpression of a series of carcinogenic genes. It was found that the lactate production rate increased proportionally with the level of HIF-1α, for it facilitates the transcription of genes related to GLUTs and glycolytic enzymes including HK, PKM, and MCT4 [11]. HIF-1 also enhanced the PDK1 function [34].
Because neovascularization frequently occurs at the periphery of tumors rather than within tumors, larger cancer tumors incline to suffer more severe hypoxia. Therefore, the tumor size and proliferation rate can often represent the extent of hypoxia damage. CircDENND4C, which was found to be upregulated by HIF-1α in breast cancer cells, aggressively promotes cell proliferation in a hypoxic environment [17]. Knowing that a larger mass consumes more energy and bears more extreme hypoxia, circDENND4C is supposed to enhance the Warburg effect. In human umbilical vascular endothelial cells (HUVECs), circ_0010729 coexists with HIF-1, especially in hypoxia-induced HUVECs, where circ_0010729 overexpressed and elevated the HIF-1α expression by sponging miR-186 [35]. With the support of thick vessels, more glucose is brought inside tumors. So circ_0010729 may also facilitate glycolysis by enhancing the progress of vascular endothelial cells.
C-myc
c-myc, one of the most important oncogenic TFs, can also activate the Warburg effect. Ectopic c-myc can collaborate with HIF-1 to elevate the level of the glycolytic components GLUT1, HK2, and PDK1 [34, 36].
In bladder cancer, miR-145 could alleviate the Warburg effect by silencing c-myc [37]. To explain detailedly, polypyrimidine tract-binding protein1 (PTBP1) as the downstream of c-myc induces the up-regulation of PKM2 instead of PKM1. Meanwhile, miR-145 can counteract the function of the c-myc/PTBP1 axis to reduce the PKM2/PKM1 ratio [38]. Two circRNAs interplay with miR-145. One is circRNA_001569 sponging miR-145 in colorectal cancer, and the other is circBIRC6 [18, 19]. circBIRC6 also targets miR-34a, another miRNA repressing c-myc [19]. The above-mentioned Circ-Amotl1 can also assist c-myc in nuclear translocation in order to prevent the degradation of c-myc [15]. Besides c-myc, circ-Amotl1 could link with classic oncogenic proteins such as AKT and STAT3. Consequently, it is highly feasible that circ-Amotl1 enhances aerobic glycolysis to facilitate tumor development without working as a competing endogenous RNA (ceRNA). Intriguingly, circ-FBXW7 could encode protein FBXW7-185aa which was capable to competitively interact with the de-ubiquitinating enzyme USP28 [20]. So FBXW7-185aa could indirectly degrade c-myc. Circ-FBXW7 probably ameliorates the Warburg effect in this way.
circRNAs affect glycolysis by regulating signaling pathways
Effective TFs work together to form a regulatory network that affects the Warburg effect. The network consists of different signaling pathways in which diverse circRNAs are involved. So reviewing the representative signaling pathways and associated circRNAs can help understand this metabolic mechanism more comprehensively.
PI3K/Akt
The PI3K/Akt pathway plays a crucial role in motivating glucose uptake and glycogen synthesis. Akt can encourage HK2 and PFK2 to stimulate glycolysis [39]. MTOR complex1, as the downstream of Akt, can phosphorylate elf4E-binding proteins and support HIF-1α to promote GLUT1, HK2, and PFK2 [40, 41].
First of all, in gastric cancer, circNRIP1 could upregulate glucose uptake and induce lactification via sponging the miR149-5p that targets AKT1 [21]. Second, it is possible that miR-150 directly depresses the Warburg effect. Downregulation of miR-150 reinforced the resistance of ovarian cancer cells to pertuzumab by increasing p-Akt [22]. And in osteosarcoma, miR-150 might repress glycolysis by targeting the 3′-UTR of GLUT1 [42]. In Hirschsprung disease, the level of AKT3 scaled up with that of circ-ZNF609, while miR-150-5p expression was inversely associated with circ-ZNF609 and AKT3 [43]. Some experimental studies found circ-ZNF609 could harbor the target sites of miR-150-5p. So circ-ZNF609 might promote AKT3 by targeting miR-150-5p to improve the Warburg effect. More research is required to confirm its role in cancer metabolism. In osteosarcoma, circRNA_103801 was found to be associated with the HIF-1 and PI3K/AKT pathway by binding with miR-370 and miR-877 as predicted by bioinformatics tools [23]. In ovarian cancer, miR-29b targeted AKT2 and AKT3 to moderate the Warburg effect [44]. And in oral squamous cell carcinoma, circRNA_100290 sponged miR-29 family to downregulate CDK6 [24]. Similarly, miR-124 could target AKT1 and AKT2 to decrease GLUT1 and HK2 to relieve the Warburg effect in non-small cell lung cancer cells [31]. And in vascular smooth muscle cells, circWDR77 has been verified to sponge miR-124 [45].
RAS
KRAS mutations occur in a variety of cancers, especially in colorectal cancer, non-small cell lung cancer, and PCa. The alteration of KRAS upregulated GLUT1 and glycolytic enzymes to facilitate the Warburg effect [46]. In bladder cancer, ectopic circRNA-MYLK increased the expression of RAS and the downstream Raf/MEK/ERK [26]. CircRNA-MYLK can activate the vascular endothelial growth factor A to elevate the glucose brought by blood as a ceRNA for miR-29a. Additionally, depending on the ceRNA network, both lncRNA H19 and circRNA-MYLK were the ceRNA of miR-29a-3p in breast cancer [27]. And H19 could activate PKM2 and PKM2 polymers in hepatocarcinogenesis, with the upregulation of c-myc and HRAS [47]. Deductively, circRNA-MYLK might sponge miR-29a to discharge more H19 to increase PKM2. Similarly, circRNA_100290 sponged the entire miR-29 family, which is perhaps helpful for the expression of RAS.
STAT3
The STAT3 pathway gives rise to the inflammatory environment in favor of cancerogenesis. Experiments in breast cancer cells have demonstrated that STAT3 could directly upregulate the expression of HK2 [48]. The aforementioned circ-Amotl1 could communicate with STAT3 to markedly increase cell proliferation [16]. By increasing the expression of STAT3 and inducing STAT3 nuclear distribution, circ-Amotl1 enhanced STAT3 expression. In osteosarcoma, hsa_circ_0009910 indirectly facilitated STAT3 with miR-449a as a mediator [49]. Circ_0009910 could sponge miR-449a which antagonized IL6R. IL6R increased BCL-2 and STAT3 in hepatocellular carcinoma (HCC). The deregulated circ_0009910 could cut down the levels of p-STAT3 and Bcl-2 [50].
Wnt/β-catenin
The Wnt/β-catenin pathway can direct the Warburg effect by upregulating the transcription of MCT1 and PDK1 [12]. Mutant Wnt signaling can activate β-catenin leading to the overactive response of the downstream genes. Furthermore, c-myc has been ascertained as a Wnt/β-catenin target [28]. In papillary thyroid cancer, circ-ITCH could sponge miR-22-3p to upregulate CBL, an E3 ligase of nuclear β-catenin [51]. CBL, which is detrimental to tumor growth and development, could degrade β-catenin. So circ-ITCH might suppress the Warburg effect by impairing the activation of the Wnt/β-catenin pathway. Additionally, circRNA_102171, circ_0067934, and circRNA_100290 were in favor of the Wnt/β-catenin pathway by targeting CTNNBIP1, miR-1324, and miR-516b, respectively [25, 52, 53], while circ_0006427 inactivated the Wnt/β-catenin pathway by targeting miR-6783–3p [54].
circRNAs in lipid metabolism
Exacerbated lipid production is another hallmark of tumor metabolism, for cancer cell growth requires fatty acids (FAs) to replicate cellular membranes. Endogenous fatty acid synthesis (FAS) is triggered in various tumors [55]. Acetyl-COA can be converted from citrate catalyzed by ATP citrate lyase. The first step of FA biosynthesis is the transformation of cytosolic acetyl-COA to malonyl-COA via acetyl-COA carboxylase. Then, fatty acid synthase (FASN) catalyses malonyl-COA into long FA chains [2]. In contrast, altered lipid mobilization also has relevance with tumor progression. For example, increased lipolysis leads to a wasting syndrome known as cancer cachexia, which is characterized by acute fat loss [56]. Tumor cells can acquire FAs through lipolysis to perform FA β-oxidation (FAO) which provides energy for tumor proliferation. Due to the Warburg effect, less acetyl-COA is generated by glucose, while FAO can comprise this deficiency. Moreover, lipophagy, meaning autophagic degradation of lipid, is another pathway of lipid utilization [57]. Recent studies indicate that lipophagy is a well-conserved mechanism of lipid degradation. Lysosomes can digest lipid droplets with the help of phagophore to reproduce free FAs [58].
An inverse correlation between circRNA_0046367 expression and triglyceride (TG) level in HepG2 cell culture and liver tissues has been reported [59]. CircRNA_0046367 could sponge miR-34a to protect the peroxisome proliferator-activated receptor (PPAR) α from transcriptional repression. PPARα activates CPT2 and ACBD3 to degrade lipids [60]. So circRNA_0046367 might promote tumor progression partly through upregulating FAO. CircGFRA1 attracted miR-34a in triple negative breast cancer as a ceRNA to release the restrain of GFRA1, which can induce autophagy [61, 62]. More studies are required to confirm whether circGFRA1 concomitant with GFRA1 can promote lipophagy in cancer metabolism. On the contrary, circ-DB released by adipocyte exosomes also sponged miR-34 but appeared to raise the body fat ratio in HCC patients [63]. HCC patients with the higher body fat ratio incline to have more rapid tumor progression and poorer prognosis. In this circumstance, circ-DB seems to facilitate HCC progression via FAS. However, this mechanism is paradoxical to circRNA_0046367 whose target is miR-34a as well. Considering that the expression of miR-34a is increased in obesity causing the fat browning and the weight loss, the difference of circ-DB function may be caused by lipid in exosomes [64]. But the authentic mechanism remains elusive. After high fat stimulation, circRNA_021412 was downregulated in HepG2 cells causing reactivation of miR-1972 [65]. Increased miR-1972 reduced the level of LPIN1, which can coordinate with PPARα to recruit long-chain acyl-CoA synthetases. The inhibition of LPIN1 blocks FAO resulting in hepatic steatosis. The circRNA_021412/miR-1972/LPIN1 axis may also be involved in tumor progression. The expression of circ_0057558 was found to be positively associated with the level of TG in PCa [66]. PCa is a hormone-dependent cancer influenced by lipid metabolism abnormalities [67]. So circ_0057558 might regulate the level of lipid to promote PCa progression. CircFARSA, which was found to be highly expressed in lung adenocarcinoma, is supposed to interact with FASN through miR-330-5p and miR-326 in accordance with the in silico analysis [68]. And in laryngeal squamous cell carcinoma, hsa_circ_0033988 was lowly expressed and associated with FA degradation according to ceRNA network analysis and functional annotation analysis [69]. These results computed by special algorithms have yet to be further validated by experiments in vitro and in vivo (Table 2).
circRNAs in amino acid metabolism
Since the 1950s, scientists have found that tumor cells consume much more glutamine than any other amino acid. Glutamine can be converted to glutamate catalyzed by glutaminase. Subsequently, glutamate can provide nitrogen for purine and pyrimidine biosynthesis, or participate in the TCA cycle by transforming to α-ketoglutarate. Glutathione, an important cellular antioxidant, also comes from glutamate. Thus, glutamine plays an essential role in tumor progression.
Despite the deficiency of direct evidence, scientists have investigated the impact of circRNAs on glutamine metabolism by comprehensive analysis (Table 3). In glioblastoma, functional analysis predicted that circRNAs may participate in glutamergic synapse and calcium signaling [70]. Computational analysis discovered the circRNA_002581/miR-122/Slc1a5 axis in non-alcoholic steatohepatitis [70]. Knowing that Slc1a5 is a kind of glutamine transporter, biological research is needed to evaluate the relationship between circRNA_002581 and glutamine metabolism [71].
circRNAs in oxidative respiration
Owing to the electron transport flux accumulated by proliferating cells and hypoxia in the tumor microenvironment, the constant generation of reactive oxygen species (ROS) is another attribute of cancer. Overexpressed ROS can develop into oxidative stress, eventually resulting in oncogene-induced cellular senescence. Although ROS overproduction is harmful for cell growth, a moderate amount of ROS is beneficial for the maintenance of a tumorigenic condition [72]. The tolerable level of ROS not only activates HIF1-α and NRF2 but inhibits protein phosphatases such as PTEN [2]. So reduction-oxidation (redox) homeostasis is a crucial component of cancer metabolism. Some studies demonstrated that several circRNAs were differentially expressed in substantia nigra and corpus striatum of NRF2 knock-out mice compared with the control group [73]. Increases CircNCX1 in response to ROS in cardiomyocytes could sponge miR-133a-3p to release the pro-apoptotic gene CDIP1 [74]. These circRNAs implicated with ROS may also regulate the redox equilibrium in tumor cells [Table 4].
The folate cycle plays a dominant role in the generation of NADPH, which effectively antagonizes ROS. Recent studies point out that folate is required for HCC and PCa cells growth [75, 76]. In PCa, circ_0062019 and its host gene SLC19A1 were significantly upregulated [66]. SLC19A1 can encode a membrane protein to transport folate. This result suggests that circ_0062019 may act as a booster to further PCa proliferation by the folate cycle.
Inductive analysis
Major regulatory mechanisms of circRNAs can be divided into three aspects. Firstly, circRNAs with miRNA response elements can serve as ceRNAs to sponge miRNAs [6]. As circRNAs lack poly (A) tails and 5′ ends, it is difficult for them to get degraded. Thereby, a small portion of circRNAs can repress a large number of miRNAs [77]. Secondly, it can interact with RNA polymerase II or splicing factors to regulate gene transcription [78]. Finally, they can combine with RBPs to compete with RBP substrates. According to the up-to-date records, we find that most circRNAs regulate cancer metabolism through sponging miRNAs. In addition, circ-FBXW7 as a substitution of its linear counterpart attracted USP28. Circ-Amotl1 induced the nuclear translocation of PDK1, AKT1, and c-mcy. These special mechanisms broaden the horizon on the research of circRNAs and might bridge to novel regulatory systems.
As a core hallmark of cancer, altered energy metabolism has its particularity and university. On the one hand, metabolism consists of distinctive chemicals, transporters, and enzymes, running for supplying necessary nutrients. On the other hand, deregulating cellular energy metabolism is also supported by TFs and ncRNAs that are implicated in other core hallmarks of cancer, such as proliferation and metastasis. To some extent, altered energy metabolism is simply another profile of oncogene programming. Therefore, for one thing, we gather the circRNAs that directly regulate enzymes, like circHIPK3 that affects GLUT1. For another, we record the circRNAs related to the TFs and pathways which assist in the formation of abnormal metabolic process, such as circ-Amotl1 and circRNA_100290. The results observably ascertain that circRNAs are of importance in regulating cellular metabolism.
Since 2015, scientists have noticed that circRNAs are enriched in exosomes compared with parental cells, and many investigators have concentrated on the relationship between circRNAs and exosomes [79]. Exosomes also contribute to the mutant metabolism in cancer. They bring the necessary nutrients for tumor growth and transport ncRNAs to regulate the signaling pathways. It is striking that adding exosomes into prostate or pancreatic cancer could promote glycolysis and reduce OXPHOS [80]. However, the underlying mechanism remains to be clarified. According to our collection of metabolic circRNAs, it is possible that exosomes apply circRNAs for the regulation of metabolism.
The metabolic phenotype of cancer stem cells (CSC) has been intensely investigated over the past years. It was reported that high levels of glucose uptake interfered with the natural differentiation of stem cells [81]. And constant stem cell metabolism lays the ground for the Warburg effect. It seems that cellular metabolism controls the “stemness” properties. Publications have reported that circRNAs are implicated with CSCs. For example, circRNA VRK1 was found to be negatively related with breast cancer stem cells [82]. CircBIRC6 could maintain stem cell pluripotency and repress differentiation by binding to miR-34 and miR-145 in human embryonic stem cells (hESCs) [19]. CircBIRC6 also has binding sites for several other miRNAs associated with primary lineage differentiation, including let-7, miR-92, and miR-103. The epithelial-splicing regulatory protein 1 (ESRP1), regulated by pluripotency-associated genes NANOG and OCT4, is responsible for circBIRC6 biosynthesis in hESCs. Therefore, circBIRC6 is probably able to induce the Warburg effect by inhibiting the differentiation of cancer cells. Moreover, FAO plays an indispensable role in CSC growth. Hematopoietic stem cells and leukemia-initiating cells appear to rely on FAO to complete self-renewal [83, 84]. This finding suggests that circRNAs implicating in FAO may regulate CSCs.
One challenge in the study of circRNAs related to cancer metabolism is the lack of direct evidence, and therefore further investigation and validation are warranted in this field. For one thing, although advanced computational models point out several circRNA-miRNA axes, few of them have been verified by experiments in vitro and in vivo. For another, we hypothesize that circRNAs might be involved in cancer metabolism by regulating special TFs directly or indirectly. But the effects of altered TFs might be compensated by corresponding downstream feedback because of complex intracellular regulatory mechanisms. Measurements of metabolic levels before and after circRNA overexpression or knockdown are inevitable. Furthermore, vital oncogenic TFs are involved in more than one metabolic pathway. For example, c-myc can promote both glycolysis and glutamine metabolism. Resultantly, circRNAs may not implicate merely one pathway. Finally, several laboratories have identified that circRNAs play a regulatory role in multiple metabolic diseases, though no sufficient evidence is available to explain their functions in cancer metabolism.
Irrespective of these defects, circRNAs are qualified for a potential therapy to precisely regulate metabolism due to their extremely high stability, strong evolutionary conservation, and unique temporal and spatial expression [77, 85]. With more ncRNAs emerging as important players of metabolism regulation, circRNAs may be able to work as a cut-in point of the whole ncRNA network and bring a promising prospect for the clinical treatment of cancers.
Conclusion
The purpose of this review is to explore circRNAs that play major roles in cancer metabolism. We have found that several circRNAs are closely associated with carbohydrate, lipid, and amino acid metabolisms. By targeting miRNAs or proteins, circRNAs work as part of the ncRNA regulatory network. In addition, circRNAs may affect exosomes or CSCs to show broad effects. The knowledge about circRNAs may help better understand altered energy metabolism in cancer cells. We envision that with the development of biological research, circRNAs may be utilized for regulating metabolisms in clinical practice in near future.
Availability of data and materials
Not applicable
Abbreviations
- ceRNA:
-
Competing endogenous RNA
- circRNA:
-
Circular RNA
- CSC:
-
Cancer stem cell
- ESRP1:
-
Epithelial-splicing regulatory protein 1
- FA:
-
Fatty acid
- FAO:
-
Fatty acid β-oxidation
- FAS:
-
Fatty acid synthesis
- FASN:
-
Fatty acid synthase
- GLUT:
-
Glucose transporter
- HCC:
-
Hepatocellular carcinoma
- hESCs:
-
Human embryonic stem cells
- HK:
-
Hexokinase
- HUVEC:
-
Human umbilical vascular endothelial cell
- LDHA:
-
Lactate dehydrogenase A
- MCT:
-
Monocarboxylate transporter
- NADPH:
-
Nicotinamide adenine dinucleotide phosphate
- ncRNA:
-
Non-coding RNA
- OXPHOS:
-
Oxidative phosphorylation
- PCa:
-
Prostate cnacer
- PDH:
-
Pyruvate dehydrogenase
- PDK:
-
Pyruvate dehydrogenase kinase
- PET:
-
Positron emission tomography
- PFK:
-
6-Phosphfructa-1-kinase
- PK:
-
Pyruvate kinase
- PTBP1:
-
Polypyrimidine tract-binding protein 1
- PPAR:
-
Peroxisome proliferator-activated receptor
- RBP:
-
RNA-binding protein
- Redox:
-
Reduction-oxidation
- ROS:
-
Reactive oxygen species
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47.
Chan B, Manley J, Lee J, Singh SR. The emerging roles of microRNAs in cancer metabolism. Cancer Lett. 2015;356(2 Pt A):301-8.
Xiao ZD, Zhuang L, Gan B. Long non-coding RNAs in cancer metabolism. Bioessays. 2016;38(10):991–6.
Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–61.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8.
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21 e7.
Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–14.
DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148(6):1132–44.
Stubbs M, Griffiths JR. The altered metabolism of tumors: HIF-1 and its role in the Warburg effect. Adv Enzyme Regul. 2010;50(1):44–55.
Pate KT, Stringari C, Sprowl-Tanio S, Wang K, TeSlaa T, Hoverter NP, et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J. 2014;33(13):1454–73.
Stoll L, Sobel J, Rodriguez-Trejo A, Guay C, Lee K, Veno MT, et al. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol Metab. 2018;9:69–83.
Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, et al. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics. 2017;7(16):3842–55.
Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24(9):1609–20.
Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J. Wu, et al. The Circular RNA Interacts with STAT3, Increasing Its Nuclear Translocation and Wound Repair by Modulating Dnmt3a and miR-17 Function. Mol Ther. 2017;25(9):2062–74.
Liang G, Liu Z, Tan L, Su AN, Jiang WG, Gong C. HIF1α-associated circDENND4C Promotes Proliferation of Breast Cancer Cells in Hypoxic Environment. Anticancer Res. 2017;37(8):4337–43.
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–91.
Yu CY, Li TC, Wu YY, Yeh CH, Chiang W, Chuang CY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8(1):1149.
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst. 2018;110(3).
Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20.
Wuerkenbieke D, Wang J, Li Y, Ma C. miRNA-150 downregulation promotes pertuzumab resistance in ovarian cancer cells via AKT activation. Arch Gynecol Obstet. 2015;292(5):1109–16.
Liu W, Zhang J, Zou C, Xie X, Wang Y, Wang B, et al. Microarray expression profile and functional analysis of circular RNAs in osteosarcoma. Cell Physiol Biochem. 2017;43(3):969–85.
Chen L, Zhang S, Wu J, Cui J, Zhong L, Zeng L, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551–61.
Fang G, Ye BL, Hu BR, Ruan XJ, Shi YX. CircRNA_100290 promotes colorectal cancer progression through miR-516b-induced downregulation of FZD4 expression and Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2018;504(1):184–9.
Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–17.
Huang M, Zhong Z, Lv M, Shu J, Tian Q, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7(30):47186–200.
Rennoll S, Yochum G. Regulation of MYC gene expression by aberrant Wnt/beta-catenin signaling in colorectal cancer. World J Biol Chem. 2015;6(4):290–300.
Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3(3):177–85.
Murphy ST, Alton G, Bailey S, Baxi SM, Burke BJ, Chappie TA, et al. Discovery of novel, potent, and selective inhibitors of 3-phosphoinositide-dependent kinase (PDK1). J Med Chem. 2011;54(24):8490–500.
Zhao X, Lu C, Chu W, Zhang B, Zhen Q, Wang R, et al. MicroRNA-124 suppresses proliferation and glycolysis in non-small cell lung cancer cells by targeting AKT-GLUT1/HKII. Tumour Biol. 2017;39(5):1010428317706215.
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215.
Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53(1):88–100.
Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27(21):7381–93.
Dang RY, Liu FL, Li Y. Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1alpha axis. Biochem Biophys Res Commun. 2017;490(2):104–10.
Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci. 2016;73(2):377–92.
Wang F, Xia J, Wang N, Zong H. miR-145 inhibits proliferation and invasion of esophageal squamous cell carcinoma in part by targeting c-Myc. Onkologie. 2013;36(12):754–8.
Takai T, Yoshikawa Y, Inamoto T, Minami K, Taniguchi K, Sugito N, et al. A Novel Combination RNAi toward Warburg Effect by Replacement with miR-145 and Silencing of PTBP1 Induces Apoptotic Cell Death in Bladder Cancer Cells. Int J Mol Sci. 2017;18(1).
Robey RB, Hay N. Is Akt the "Warburg kinase"?-Akt-energy metabolism interactions and oncogenesis. Semin Cancer Biol. 2009;19(1):25–31.
Yang X, Cheng Y, Li P, Tao J, Deng X, Zhang X, et al. A lentiviral sponge for miRNA-21 diminishes aerobic glycolysis in bladder cancer T24 cells via the PTEN/PI3K/AKT/mTOR axis. Tumour Biol. 2015;36(1):383–91.
Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, et al. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339(6125):1320–3.
Yuan G, Zhao Y, Wu D, Gao C. Mir-150 Up-Regulates Glut1 and Increases Glycolysis in Osteosarcoma Cells. Asian Pac J Cancer Prev. 2017;18(4):1127–31.
Peng L, Chen G, Zhu Z, Shen Z, Du C, Zang R, et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease. Oncotarget. 2017;8(1):808–18.
Teng Y, Zhang Y, Qu K, Yang X, Fu J, Chen W, et al. MicroRNA-29B (mir-29b) regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3. Oncotarget. 2015;6(38):40799–814.
Chen J, Cui L, Yuan J, Zhang Y, Sang H. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem Biophys Res Commun. 2017;494(1-2):126–32.
Kawada K, Toda K, Sakai Y. Targeting metabolic reprogramming in KRAS-driven cancers. Int J Clin Oncol. 2017;22(4):651–9.
Li H, Li J, Jia S, Wu M, An J, Zheng Q, et al. miR675 upregulates long noncoding RNA H19 through activating EGR1 in human liver cancer. Oncotarget. 2015;6(31):31958–84.
Jiang S, Zhang LF, Zhang HW, Hu S, Lu MH, Liang S, et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012;31(8):1985–98.
Deng N, Li L, Gao J, Zhou J, Wang Y, Wang C, et al. Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem Biophys Res Commun. 2017.
Liu X, Zhang A, Xiang J, Lv Y, Zhang X. miR-451 acts as a suppressor of angiogenesis in hepatocellular carcinoma by targeting the IL-6R-STAT3 pathway. Oncol Rep. 2016;36(3):1385–92.
Wang M, Chen B, Ru Z, Cong L. CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/beta-catenin pathway. Biochem Biophys Res Commun. 2018;504(1):283–8.
Bi W, Huang J, Nie C, Liu B, He G, Han J, et al. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of beta-catenin pathway. J Exp Clin Cancer Res. 2018;37(1):275.
Zhu Q, Lu G, Luo Z, Gui F, Wu J, Zhang D, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem Biophys Res Commun. 2018;497(2):626–32.
Yao Y, Hua Q, Zhou Y. CircRNA has_circ_0006427 suppresses the progression of lung adenocarcinoma by regulating miR-6783-3p/DKK1 axis and inactivating Wnt/beta-catenin signaling pathway. Biochem Biophys Res Commun. 2019;508(1):37–45.
Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7(10):763–77.
Gomez de Cedron M, Ramirez de Molina A. Microtargeting cancer metabolism: opening new therapeutic windows based on lipid metabolism. J Lipid Res. 2016;57(2):193–206.
Maan M, Peters JM, Dutta M, Patterson AD. Lipid metabolism and lipophagy in cancer. Biochem Biophys Res Commun. 2018;504(3):582–9.
Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–5.
Guo XY, Chen JN, Sun F, Wang YQ, Pan Q, Fan JG. circRNA_0046367 prevents hepatoxicity of lipid peroxidation: an inhibitory role against hepatic steatosis. Oxid Med Cell Longev. 2017;2017:3960197.
Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J Clin Invest. 2006;116(3):571–80.
Kim M, Jung JY, Choi S, Lee H, Morales LD, Koh JT, et al. GFRA1 promotes cisplatin-induced chemoresistance in osteosarcoma by inducing autophagy. Autophagy. 2017;13(1):149–68.
He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36(1):145.
Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2018.
Yang Z, Cappello T, Wang L. Emerging role of microRNAs in lipid metabolism. Acta Pharm Sin B. 2015;5(2):145–50.
Guo XY, He CX, Wang YQ, Sun C, Li GM, Su Q, et al. Circular RNA profiling and bioinformatic modeling identify its regulatory role in hepatic steatosis. Biomed Res Int. 2017;2017:5936171.
Xia Q, Ding T, Zhang G, Li Z, Zeng L, Zhu Y, et al. Circular RNA expression profiling identifies prostate cancer-specific circRNAs in prostate cancer. Cell Physiol Biochem. 2018;50(5):1903-1915.
Deep G, Schlaepfer IR. Aberrant lipid metabolism promotes prostate cancer: role in cell survival under hypoxia and extracellular vesicles biogenesis. Int J Mol Sci. 2016;17(7).
Hang D, Zhou J, Qin N, Zhou W, Ma H, Jin G, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7(6):2783–91.
Zhao R, Li FQ, Tian LL, Shang DS, Guo Y, Zhang JR, et al. Comprehensive analysis of the whole coding and non-coding RNA transcriptome expression profiles and construction of the circRNA-lncRNA co-regulated ceRNA network in laryngeal squamous cell carcinoma. Funct Integr Genomics. 2019;19(1):109–21.
Zhang F, Zhang R, Zhang X, Wu Y, Li X, Zhang S, et al. Comprehensive analysis of circRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of atherosclerosis in rabbits. Aging (Albany NY). 2018;10(9):2266–83.
Bode BP, Fuchs BC, Hurley BP, Conroy JL, Suetterlin JE, Tanabe KK, et al. Molecular and functional analysis of glutamine uptake in human hepatoma and liver-derived cells. Am J Physiol Gastrointest Liver Physiol. 2002;283(5):G1062–73.
Sullivan LB, Chandel NS. Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2014;2:17.
Yang JH, Zhang RJ, Lin JJ, Cao MC, Wang Q, Cui HX, et al. The differentially expressed circular RNAs in the substantia nigra and corpus striatum of Nrf2-knockout mice. Cell Physiol Biochem. 2018;50(3):936–51.
Li M, Ding W, Tariq MA, Chang W, Zhang X, Xu W, et al. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics. 2018;8(21):5855–69.
Lee D, Xu IM, Chiu DK, Lai RK, Tse AP, Lan Li L, et al. Folate cycle enzyme MTHFD1L confers metabolic advantages in hepatocellular carcinoma. J Clin Invest. 2017;127(5):1856–72.
Tomaszewski JJ, Cummings JL, Parwani AV, Dhir R, Mason JB, Nelson JB, et al. Increased cancer cell proliferation in prostate cancer patients with high levels of serum folate. Prostate. 2011;71(12):1287–93.
Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30(21):4414–22.
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806.
Lener T, Gimona M, Aigner L, Borger V, Buzas E, Camussi G, et al. Applying extracellular vesicles based therapeutics in clinical trials - an ISEV position paper. J Extracell Vesicles. 2015;4:30087.
Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T, et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250.
Riester M, Xu Q, Moreira A, Zheng J, Michor F, Downey RJ. The Warburg effect: persistence of stem-cell metabolism in cancers as a failure of differentiation. Ann Oncol. 2018;29(1):264–70.
Yan N, Xu H, Zhang J, Xu L, Zhang Y, Zhang L, et al. Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget. 2017;8(56):95704–18.
Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest. 2010;120(1):142–56.
Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18(9):1350–8.
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–47.
Acknowledgements
Not applicable
Funding
This study was supported by the National Natural Science Foundation of China (81672896 and 81672071), the Natural Science Foundation of Jiangsu Province for Youth (BK20161066), the National Key Research and Development Program: the Key Technology of Palliative Care and Nursing for Cancer Patients (ZDZX2017ZL-01), the Jiangsu Provincial Key Research and Development Special Fund (BE2015666), the Jiangsu Innovative team leading talent fund (CXTDC2016006), Jiangsu Natural Science Foundation (BK20171304), the Natural Science Foundation of Jiangsu Provincial Department of Education (17KJB320007), and the Csco-Hawson Cancer Research Fund (Y-HS2017-032).
Author information
Authors and Affiliations
Contributions
All authors have contributed to the preparation of this manuscript. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yu, T., Wang, Y., Fan, Y. et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol 12, 90 (2019). https://doi.org/10.1186/s13045-019-0776-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13045-019-0776-8