Abstract
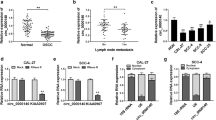
Oral squamous cell carcinoma (OSCC) is a common malignant tumor with high recurrence, metastasis rates, and poor prognosis. Numerous studies discover that circular RNA (circRNA) is closely associated with OSCC progression. Hsa_circ_0020377 has been aberrantly expressed in OSCC, but its role in tumor growth and metastasis remains largely unclear. Hsa_circ_0020377, microRNA-194-5p (miR-194-5p), and Krüppel-like factor 7 (KLF7) contents were determined by real-time quantitative polymerase chain reaction (RT-qPCR). Cell proliferative, cycle progression migration, and invasion were measured using 5-ethynyl-2′-deoxyuridine (EdU), Cell Counting Kit-8 (CCK-8), flow cytometry, wound healing, and Transwell assays. The glycolysis level was detected via specific kits. Cyclin D1, E-cadherin, hexokinase 2 (HK2), and KLF7 protein levels were detected via western blot. Using predicting bioinformatics software, the binding between miR-194-5p and hsa_circ_0020377 or KLF7 was verified using a dual-luciferase reporter and RNA Immunoprecipitation (RIP). Beyond that, a xenograft tumor model was used to analyze the role of hsa_circ_0020377 on tumor cell growth in vivo. Increased hsa_circ_0020377 and KLF7 and reduced miR-194-5p were found in OSCC tissues and cell lines. Loss-of-function experiments proved that hsa_circ_0020377 depletion might block OSCC cell proliferation, cycle progression, migration, invasion, and glycolysis in vitro. In xenograft mouse models, hsa_circ_0020377 silencing might suppress tumor growth. In addition, mechanism research suggested that hsa_circ_0020377 could bind with miR-194-5p and enhance its target gene (KLF7), thereby affecting OSCC development. These results broaden our insights regarding the regulation of OSCC progression via circRNA and act as a reference for future clinical studies in OSCC diagnosis and treatment.








Similar content being viewed by others
Data availability
Not applicable.
References
Ahmad W, Yaqoob MA, Noor NFM, Ghazali FMM, Rahman NA, Tang L, Aleng NA, Alam MK (2021) The predictive model of oral squamous cell survival carcinoma: a methodology of validation. Biomed Res Int 2021:5436894
Ai Y, Wu S, Zou C, Wei H (2022) Circular RNA circFOXO3 regulates KDM2A by targeting miR-214 to promote tumor growth and metastasis in oral squamous cell carcinoma. J Cell Mol Med 26(6):1842–1852
Ala U (2020) Competing endogenous RNAs, non-coding RNAs and diseases: an intertwined story. Cells 9(7):1574
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233
Chamoli A, Gosavi AS, Shirwadkar UP, Wangdale KV, Behera SK, Kurrey NK, Kalia K, Mandoli A (2021) Overview of oral cavity squamous cell carcinoma: risk factors, mechanisms, and diagnostics. Oral Oncol 121:105451
Chen LL (2020) The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 21(8):475–490
Chen X, Yu J, Tian H, Shan Z, Liu W, Pan Z, Ren J (2019) Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-378a to regulate oral squamous cell carcinoma cells growth via glucose transporter-1 (GLUT1) and glycolysis. J Cell Physiol 234(11):19130–19140
Chen M, Zhang R, Lu L, Du J, Chen C, Ding K, Wei X, Zhang G, Huang Y, Hou J (2020) LncRNA PVT1 accelerates malignant phenotypes of bladder cancer cells by modulating miR-194-5p/BCLAF1 axis as a ceRNA. Aging 12(21):22291–22312
Chen L, Wang C, Sun H, Wang J, Liang Y, Wang Y, Wong G (2021) The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform 22(2):1706–1728
Chi H (2015) miR-194 regulated AGK and inhibited cell proliferation of oral squamous cell carcinoma by reducing PI3K-Akt-FoxO3a signaling. Biomed Pharmacother 71:53–57
Cocquerelle C, Mascrez B, Hetuin D, Bailleul B (1993) Mis-splicing yields circular RNA molecules. FASEB J 7(1):155–160
Cui X, Wang J, Guo Z, Li M, Li M, Liu S, Liu H, Li W, Yin X, Tao J et al (2018) Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer 17(1):123
Ding X, Wang X, Gong Y, Ruan H, Sun Y, Yu Y (2017) KLF7 overexpression in human oral squamous cell carcinoma promotes migration and epithelial-mesenchymal transition. Oncol Lett 13(4):2281–2289
Fan HY, Jiang J, Tang YJ, Liang XH, Tang YL (2020) CircRNAs: a new chapter in oral squamous cell carcinoma biology. Onco Targets Ther 13:9071–9083
Glenfield C, McLysaght A (2018) Pseudogenes provide evolutionary evidence for the competitive endogenous RNA hypothesis. Mol Biol Evol 35(12):2886–2899
Guo Y, Chai B, Jia J, Yang M, Li Y, Zhang R, Wang S, Xu J (2021) KLF7/VPS35 axis contributes to hepatocellular carcinoma progression through CCDC85C-activated β-catenin pathway. Cell Biosci 11(1):73
Gupta R, Malvi P, Parajuli KR, Janostiak R, Bugide S, Cai G, Zhu LJ, Green MR, Wajapeyee N (2020) KLF7 promotes pancreatic cancer growth and metastasis by up-regulating ISG expression and maintaining Golgi complex integrity. Proc Natl Acad Sci USA 117(22):12341–12351
He Y, Deng F, Zhao S, Zhong S, Zhao J, Wang D, Chen X, Zhang J, Hou J, Zhang W et al (2019) Analysis of miRNA-mRNA network reveals miR-140-5p as a suppressor of breast cancer glycolysis via targeting GLUT1. Epigenomics 11(9):1021–1036
Hombach S, Kretz M (2016) Non-coding RNAs: classification, biology and functioning. Adv Exp Med Biol 937:3–17
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32(5):453–461
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20(11):675–691
Li J, Sun D, Pu W, Wang J, Peng Y (2020) Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer 6(4):319–336
Li Y, Gong L, Qin N, Wei X, Miao L, Yuan H, Wang C, Ma H (2021a) Comprehensive analysis of circRNA expression pattern and circRNA-miRNA-mRNA network in oral squamous cell carcinoma. Oral Oncol 121:105437
Li H, Shen H, Xie P, Zhang Z, Wang L, Yang Y, Yu Z, Cheng Z, Zhou J (2021b) Role of long intergenic non-protein coding RNA 00152 in pancreatic cancer glycolysis via the manipulation of the microRNA-185-5p/Kruppel-like factor 7 axis. J Cancer 12(21):6330–6343
Li Y, Wang Q, Wang D, Fu W (2021c) KLF7 promotes gastric carcinogenesis through regulation of ANTXR1. Cancer Manag Res 13:5547–5557
Liu J, Li D, Luo H, Zhu X (2019) Circular RNAs: the star molecules in cancer. Mol Aspects Med 70:141–152
Liu J, Jiang X, Zou A, Mai Z, Huang Z, Sun L, Zhao J (2021) circIGHG-induced epithelial-tomMesenchymal transition promotes oral squamous cell carcinoma Progression via miR-142-5p/IGF2BP3 signaling. Can Res 81(2):344–355
López-Jiménez E, Rojas AM, Andrés-León E (2018) RNA sequencing and prediction tools for circular RNAs analysis. Adv Exp Med Biol 1087:17–33
Lyu D, Huang S (2017) The emerging role and clinical implication of human exonic circular RNA. RNA Biol 14(8):1000–1006
Lyu J, Wang J, Miao Y, Xu T, Zhao W, Bao T, Zhu H (2022) KLF7 is associated with poor prognosis and regulates migration and adhesion in tongue cancer. Oral Dis 28(3):577–584
Munkley J, Elliott DJ (2016) Hallmarks of glycosylation in cancer. Oncotarget 7(23):35478–35489
Nguyen CTK, Sawangarun W, Mandasari M, Morita KI, Harada H, Kayamori K, Yamaguchi A, Sakamoto K (2020) AIRE is induced in oral squamous cell carcinoma and promotes cancer gene expression. PLoS ONE 15(2):e0222689
Panarese I, Aquino G, Ronchi A, Longo F, Montella M, Cozzolino I, Roccuzzo G, Colella G, Caraglia M, Franco R (2019) Oral and oropharyngeal squamous cell carcinoma: prognostic and predictive parameters in the etiopathogenetic route. Expert Rev Anticancer Ther 19(2):105–119
Panni S, Lovering RC, Porras P, Orchard S (2020) Non-coding RNA regulatory networks. Biochim Biophys Acta 1863(6):194417
Patop IL, Wust S, Kadener S (2019) Past, present, and future of circRNAs. EMBO J 38(16):e100836
Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W (2015) ceRNA in cancer: possible functions and clinical implications. J Med Genet 52(10):710–718
Shanti RM, Tanaka T, Stanton DC (2020) Oral biopsy techniques. Dermatol Clin 38(4):421–427
Siegel RL, Miller KD, Fuchs HE, Jemal A (2022) Cancer statistics, 2022. CA: Cancer J Clin 72(1):7–33
Swamynathan SK (2010) Kruppel-like factors: three fingers in control. Hum Genomics 4(4):263–270
Tan X, Zhou C, Liang Y, Lai YF, Liang Y (2020) Circ_0001971 regulates oral squamous cell carcinoma progression and chemosensitivity by targeting miR-194/miR-204 in vitro and in vivo. Eur Rev Med Pharmacol Sci 24(5):2470–2481
Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F et al (2011) Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147(2):344–357
Urashima M, Hama T, Suda T, Suzuki Y, Ikegami M, Sakanashi C, Akutsu T, Amagaya S, Horiuchi K, Imai Y et al (2013) Distinct effects of alcohol consumption and smoking on genetic alterations in head and neck carcinoma. PLoS ONE 8(11):e80828
Vander Heiden MG, DeBerardinis RJ (2017) Understanding the Intersections between metabolism and cancer biology. Cell 168(4):657–669
Wang Y, Sun G, Wang C, Guo W, Tang Q, Wang M (2018) MiR-194-5p inhibits cell migration and invasion in bladder cancer by targeting E2F3. J BUON 23(5):1492–1499
Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu Q, Tong X, Yang W, Xu Q, Huang D et al (2019) A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer 18(1):28
Wang F, Ji X, Wang J, Ma X, Yang Y, Zuo J, Cui J (2020) LncRNA PVT1 enhances proliferation and cisplatin resistance via regulating miR-194-5p/HIF1a axis in oral squamous cell carcinoma. Onco Targets Ther 13:243–252
Wang C, Liu WR, Tan S, Zhou JK, Xu X, Ming Y, Cheng J, Li J, Zeng Z, Zuo Y et al (2022) Characterization of distinct circular RNA signatures in solid tumors. Mol Cancer 21(1):63
Yen YT, Yang JC, Chang JB, Tsai SC (2021) Down-regulation of miR-194–5p for predicting metastasis in breast cancer cells. Int J Mol Sci 23(1):325
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, Shu Y (2019) CircRNAs in cancer metabolism: a review. J Hematol Oncol 12(1):90
Zhang Z, Yang T, Xiao J (2018) Circular RNAs: promising biomarkers for human diseases. EBioMedicine 34:267–274
Zhang B, Wang Z, Shen Y, Yang H (2020a) Silencing circular RNA hsa_circ_009755 promotes growth and metastasis of oral squamous cell carcinoma. Genomics 112(6):5275–5281
Zhang Z, Gao Z, Rajthala S, Sapkota D, Dongre H, Parajuli H, Suliman S, Das R, Li L, Bindoff LA et al (2020b) Metabolic reprogramming of normal oral fibroblasts correlated with increased glycolytic metabolism of oral squamous cell carcinoma and precedes their activation into carcinoma associated fibroblasts. Cell Mol Life Sci 77(6):1115–1133
Zheng Z, Ma X, Li H (2020) Circular RNA circMDM2 accelerates the glycolysis of oral squamous cell carcinoma by targeting miR-532-3p/HK2. J Cell Mol Med 24(13):7531–7537
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y et al (2018) Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 17(1):79
Funding
This study was supported by the Hebei Provincial Health Department Key Science and Technology Research Plan (No. 20201111) and the Youth Science and technology Project of Hebei Provincial Department of Health (No. 20190694).
Author information
Authors and Affiliations
Contributions
Peng Liu designed and performed the research; Linyu Jin, Shixiong Peng, and Yang Bao analyzed the data; NaiHeng Hei wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Written informed consents were obtained from all participants and this study was permitted by the Ethics Committee of the fourth hospital of Hebei Medical University. Consent for publication is not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

Fig. S1
Transfection efficiency of hsa_circ_0020377, miR-194-5p, and KLF7 in OSCC cell lines. (A) RT-qPCR analysis of hsa_circ_0020377 expression in SCC-9 and HSC-3 cells transfected with sh-NC or sh-hsa_circ_0020377. (B) miR-194-5p content was detected in SCC-9 and HSC-3 cells transfected with miR-NC or miR-194-5p using RT-qPCR. (C) Western blot analysis of KLF7 protein level in SCC-9 and HSC-3 cells transfected vector or KLF7. *P <0.05. (PNG 151 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hei, N., Liu, P., Jin, L. et al. Circular hsa_circ_0020377 regulates KLF7 by targeting miR-194-5p to facilitate tumor cell malignant behaviors and glycolysis in oral squamous cell carcinoma progression. Funct Integr Genomics 23, 52 (2023). https://doi.org/10.1007/s10142-023-00973-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-023-00973-w