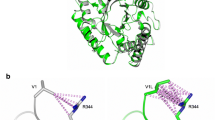
Abstract
XynII is a family 11 glycoside hydrolase that uses the retaining mechanism for catalysis. In the active site, E177 works as the acid/base and E86 works as the nucleophile. Mutating an uncharged residue (N44) to an acidic residue (D) near E177 decreases the enzyme’s optimal pH by ~ 1.0 unit. D44 was previously suggested to be a second proton carrier for catalysis. To test this hypothesis, we abolished the activity of E177 by mutating it to be Q, and mutated N44 to be D or E. These double mutants have dramatically decreased activities. Our high-resolution crystallographic structures and the microscopic pKa calculations show that D44 has similar position and pKa value during catalysis, indicating that D44 changes electrostatics around E177, which makes it prone to rotate as the acid/base in acidic conditions, thus decreases the pH optimum. Our results could be helpful to design enzymes with different pH optimum.




Similar content being viewed by others
References
Payne CM, Knott BC, Mayes HB, Hansson H, Himmel ME, Sandgren M, Stahlberg J, Beckham GT (2015) Fungal Cellulases. Chem Rev 115:1308–1448
Speciale G, Thompson AJ, Davies GJ, Williams SJ (2014) Dissecting conformational contributions to glycosidase catalysis and inhibition. Curr Opin Struc Biol 28:1–13
Langan P, Gnanakaran S, Rector KD, Pawley N, Fox DT, Cho DW, Hammel KE (2011) Exploring new strategies for cellulosic biofuels production. Energ Environ Sci 4:3820–3833
Torronen A, Harkki A, Rouvinen J (1994) Three-dimensional structure of endo-1,4-beta-xylanase II from Trichoderma reesei: two conformational states in the active site. EMBO J 13:2493–2501
Wan Q, Zhang Q, Hamilton-Brehm S, Weiss K, Mustyakimov M, Coates L, Langan P, Graham D, Kovalevsky A (2014) X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism. Acta Crystallogr SectD Biol Crystallogr. 70:11–23
Wan Q, Parks JM, Hanson BL, Fisher SZ, Ostermann A, Schrader TE, Graham DE, Coates L, Langan P, Kovalevsky A (2015) Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography. Proc Natl Acad Sci USA 112:12384–12389
Vasella A, Davies GJ, Bohm M (2002) Glycosidase mechanisms. Curr. Opin. Chem Biol 6:619–629
Zechel DL, Withers SG (2000) Glycosidase mechanisms: Anatomy of a finely tuned catalyst. Acc Chem Res 33:11–18
Bai WQ, Cao YF, Liu J, Wang QH, Jia ZH (2016) Improvement of alkalophilicity of an alkaline xylanase Xyn11A-LC from Bacillus sp SN5 by random mutation and Glu135 saturation mutagenesis. BMC Biotechnol 16:77–86
Amel BD, Nawel B, Khelifa B, Mohammed G, Manon J, Salima KG, Farida N, Hocine H, Bernard O, Jean-Luc C, Marie-Laure F (2016) Characterization of a purified thermostable xylanase from Caldicoprobacter algeriensis sp nov strain TH7C1(T). Carbohydr Res 419:60–68
Polizeli MLTM, Rizzatti ACS, Monti R, Terenzi HF, Jorge JA, Amorim DS (2005) Xylanases from fungi: properties and industrial applications. Appl Microbiol Biotechnol 67:577–591
Fushinobu S, Ito K, Konno M, Wakagi T, Matsuzawa H (1998) Crystallographic and mutational analyses of an extremely acidophilic and acid-stable xylanase: biased distribution of acidic residues and importance of Asp37 for catalysis at low pH. Protein Eng 11:1121–1128
Joshi MD, Sidhu G, Pot I, Brayer GD, Withers SG, McIntosh LP (2000) Hydrogen bonding and catalysis: A novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase. J Mol Biol 299:255–279
Li ZH, Zhang XS, Wang QQ, Li CR, Zhang NY, Zhang XK, Xu BR, Ma BL, Schrader TE, Coates L, Kovalevsky A, Huang YD, Wan Q (2018) Understanding the pH-Dependent Reaction Mechanism of a Glycoside Hydrolase Using High-Resolution X-ray and Neutron Crystallography. ACS Catal 8:8058–8069
Bornhorst JA, Falke JJ (2000) Purification of proteins using polyhistidine affinity tags. Method Enzymol 326:245–254
Bailey MJ, B. P., Poutanen K., (1992) Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol 23:257–270
Wang QS, Zhang KH, Cui Y, Wang ZJ, Pan QY, Liu K, Sun B, Zhou H, Li MJ, Xu Q, Xu CY, Yu F, He JH (2018) Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl Sci Tech 29:68
Minor W, Cymborowski M, Otwinowski Z, Chruszcz M (2006) HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr Sect D: Biol Crystallogr 62:859–866
Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D: Biol Crystallogr 66:213–221
Watanabe N, Akiba T, Kanai R, Harata K (2006) Structure of an orthorhombic form of xylanase II from Trichoderma reesei and analysis of thermal displacement. Acta Crystallogr Sect D: Biol Crystallogr 62:784–792
Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr Sect D Biol Crystallogr 68:352–367
Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr Sect D: Biol Crystallogr 66:486–501
Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D: Biol Crystallogr 66:12–21
Jeffrey GA, Yates JH (1979) Stereographic Representation of the Cremer-Pople Ring-Puckering Parameters for Pyranoid Rings. Carbohydr Res 74:319–322
Anandakrishnan R, Aguilar B, Onufriev AV (2012) H++ 3.0 automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res 40:537–541
Zhou HX, Pang XD (2018) Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Chem Rev 118:1691–1741
Sun ZX, Wang XH, Song JN (2017) Extensive Assessment of Various Computational Methods for Aspartate’s pK(a) Shift. J Chem Inf Model 57:1621–1639
Cheng YS, Chen CC, Huang CH, Ko TP, Luo W, Huang JW, Liu JR, Guo RT (2014) Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum: insights into the molecular basis of a thermophilic enzyme. J Biol Chem 289:11020–11028
Iglesias-Fernandez J, Raich L, Ardevol A, Rovira C (2015) The complete conformational free energy landscape of beta-xylose reveals a two-fold catalytic itinerary for beta-xylanases. Chem Sci 6:1167–1177
Rye CS, Withers SG (2000) Glycosidase mechanisms. Curr Opin Chem Biol 4:573–580
Ludwiczek ML, D’Angelo I, Yalloway GN, Brockerman JA, Okon M, Nielsen JE, Strynadka NCJ, Withers SG, McIntosh LP (2013) Strategies for Modulating the pH-Dependent Activity of a Family 11 Glycoside Hydrolase. Biochemistry 52:3138–3156
Joshi MD, Sidhu G, Nielsen JE, Brayer GD, Withers SG, McIntosh LP (2001) Dissecting the electrostatic interactions and pH-dependent activity of a family 11 glycosidase. Biochemistry 40:10115–10139
Ardevol A, Rovira C (2015) Reaction Mechanisms in Carbohydrate-Active Enzymes: Glycoside Hydrolases and Glycosyltransferases. Insights from ab Initio Quantum Mechanics/Molecular Mechanics Dynamic Simulations. J Am Chem Soc 137:7528–7547
Kwon H, Langan PS, Coates L, Raven EL, Moody PCE (2018) The rise of neutron cryo-crystallography. Acta Crystallogr Sect D: Struct Biol 74:792–799
Chen JCH, Unkefer CJ (2017) Fifteen years of the Protein Crystallography Station: the coming of age of macromolecular neutron crystallography. IUCrJ 4:72–86
O’Dell WB, Bodenheimer AM, Meilleur F (2016) Neutron protein crystallography: A complementary tool for locating hydrogens in proteins. Arch Biochem Biophys 602:48–60
Henzler-Wildman K, Kern D (2007) Dynamic personalities of proteins. Nature 450:964–972
Li JH, Wang LS (2011) Why substituting the asparagine at position 35 in Bacillus circulans xylanase with an aspartic acid remarkably improves the enzymatic catalytic activity? A quantum chemistry-based calculation study. Polym Degrad Stab 96:1009–1014
Acknowledgements
Q.W. was supported by the National Natural Science Foundation of China (No. 31670790), the Fundamental Research Funds for the Central Universities (No. KYXK202009), a Chinese Spallation Neutron Source User Special Grant, the Qing Lan Project of Jiangsu Province, and the Six Talent Peaks Project of Jiangsu Province. We thank the staff of the BL17U, BL18U1 and BL19U1 beamlines at Shanghai Synchrotron Radiation Facility, Shanghai, P.R. China, for assistance during X-ray data collection. Research at ORNL’s HFIR and Spallation Neutron Source was sponsored by the Scientific User Facilities Division, Office of Basic Energy Sciences, U.S. Department of Energy.
Funding
The authors declare no competing financial interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Z., Zhang, X., Li, C. et al. Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography. Protein J 39, 671–680 (2020). https://doi.org/10.1007/s10930-020-09938-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-020-09938-5