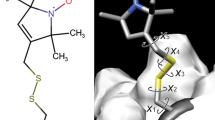
Abstract
Encodable lanthanide binding tags (LBTs) have become an attractive tool in modern structural biology as they can be expressed as fusion proteins of targets of choice. Previously, we have demonstrated the feasibility of inserting encodable LBTs into loop positions of interleukin-1β (Barthelmes et al. in J Am Chem Soc 133:808–819, 2011). Here, we investigate the differences in fast dynamics of selected loop-LBT interleukin-1β constructs by measuring 15N nuclear spin relaxation experiments. We show that the loop-LBT does not significantly alter the dynamic motions of the host protein in the sub-τc-timescale and that the loop-LBT adopts a rigid conformation with significantly reduced dynamics compared to the terminally attached encodable LBT leading to increased paramagnetic alignment strength. We further analyze residual dipolar couplings (RDCs) obtained by loop-LBTs and additional liquid crystalline media to assess the applicability of the loop-LBT approach for RDC-based methods to determine structure and dynamics of proteins, including supra-τc dynamics. Using orthogonalized linear combinations (OLCs) of RDCs and Saupe matrices, we show that the combined use of encodable LBTs and external alignment media yields up to five linear independent alignments.




Similar content being viewed by others
References
Barthelmes K, Reynolds AM, Peisach E et al (2011) Engineering encodable lanthanide-binding tags into loop regions of proteins. J Am Chem Soc 133:808–819. doi:10.1021/ja104983t
Barthelmes D, Gränz M, Barthelmes K et al (2015) Encoded loop-lanthanide-binding tags for long-range distance measurements in proteins by NMR and EPR spectroscopy. J Biomol NMR 63:275–282. doi:10.1007/s10858-015-9984-x
Bertini I, Janik MBL, Lee YM et al (2001) Magnetic susceptibility tensor anisotropies for a lanthanide ion series in a fixed protein matrix. J Am Chem Soc 123:4181–4188. doi:10.1021/ja0028626
Briggman KB, Tolman JR (2003) De novo determination of bond orientations and order parameters from residual dipolar couplings with high accuracy. J Am Chem Soc 125:10164–10165. doi:10.1021/ja035904+
Daube D, Aladin V, Heiliger J et al (2016) Heteronuclear cross-relaxation under solid-state dynamic nuclear polarization. J Am Chem Soc 138:16572–16575. doi:10.1021/jacs.6b08683
Gebel EB, Ruan K, Tolman JR, Shortle D (2006) Multiple alignment tensors from a denatured protein. J Am Chem Soc 128:9310–9311. doi:10.1021/ja0627693
Higman VA, Boyd J, Smith LJ, Redfield C (2011) Residual dipolar couplings: are multiple independent alignments always possible? J Biomol NMR 49:53–60. doi:10.1007/s10858-010-9457-1
Keizers PHJ, Ubbink M (2011) Paramagnetic tagging for protein structure and dynamics analysis. Prog Nucl Magn Reson Spectrosc 58:88–96. doi:10.1016/j.pnmrs.2010.08.001
Koehler J, Meiler J (2011) Expanding the utility of NMR restraints with paramagnetic compounds: background and practical aspects. Prog Nucl Magn Reson Spectrosc 59:360–389. doi:10.1016/j.pnmrs.2011.05.001
Lorieau JL, Maltsev AS, Louis JM, Bax A (2013) Modulating alignment of membrane proteins in liquid-crystalline and oriented gel media by changing the size and charge of phospholipid bicelles. J Biomol NMR 55:369–377. doi:10.1007/s10858-013-9720-3
Martin LJ, Hähnke MJ, Nitz M et al (2007) Double-lanthanide-binding tags: design, photophysical properties, and NMR applications. J Am Chem Soc 129:7106–7113. doi:10.1021/ja070480v
Mascali FC, Ching HYV, Rasia RM et al (2016) Using genetically encodable self-assembling Gd(III) spin labels to make in-cell nanometric distance measurements. Angew Chem Int Ed Engl 55:11041–11043. doi:10.1002/anie.201603653
Meirovitch E, Lee D, Walter KFA, Griesinger C (2012) Standard tensorial analysis of local ordering in proteins from residual dipolar couplings. J Phys Chem B 116:6106–6117. doi:10.1021/jp301451v
Musial-Siwek M, Jaffee MB, Imperiali B (2016) Probing polytopic membrane protein-substrate interactions by luminescence resonance energy transfer. J Am Chem Soc 138:3806–3812. doi:10.1021/jacs.5b13426
Peti W, Meiler J, Brüschweiler R, Griesinger C (2002) Model-free analysis of protein backbone motion from residual dipolar couplings. J Am Chem Soc 124:5822–5833. doi:10.1021/ja011883c
Reynolds AM, Sculimbrene BR, Imperiali B (2008) Lanthanide-binding tags with unnatural amino acids: Sensitizing Tb 3+ and Eu 3+ luminescence at longer wavelengths. Bioconjug Chem 19:588–591. doi:10.1021/bc700426c
Ruan K, Tolman JR (2005) Composite alignment media for the measurement of independent sets of NMR residual dipolar couplings. J Am Chem Soc 127:15032–15033. doi:10.1021/ja055520e
Ruan K, Briggman KB, Tolman JR (2008) De novo determination of internuclear vector orientations from residual dipolar couplings measured in three independent alignment media. J Biomol NMR 41:61–76. doi:10.1007/s10858-008-9240-8
Salmon L, Bouvignies G, Markwick P et al (2009) Protein conformational flexibility from structure-free analysis of NMR dipolar couplings: quantitative and absolute determination of backbone motion in ubiquitin. Angew Chem Int Ed Engl 48:4154–4157. doi:10.1002/anie.200900476
Sass J, Cordier F, Hoffmann A et al (1999) Purple membrane induced alignment of biological macromolecules in the magnetic field. J Am Chem Soc 121:2047–2055. doi:10.1021/ja983887w
Silvaggi NR, Martin LJ, Schwalbe H et al (2007) Double-lanthanide-binding tags for macromolecular crystallographic structure determination. J Am Chem Soc 129:7114–7120. doi:10.1021/ja070481n
Su XC, McAndrew K, Huber T, Otting G (2008) Lanthanide-binding peptides for NMR measurements of residual dipolar couplings and paramagnetic effects from multiple angles. J Am Chem Soc 130:1681–1687. doi:10.1021/ja076564l
Tolman JR (2002) A novel approach to the retrieval of structural and dynamic information from residual dipolar couplings using several oriented media in biomolecular NMR spectroscopy. J Am Chem Soc 124:12020–12030. doi:10.1021/ja0261123
Tolman JR (2009) NEWS & VIEWS protein dynamics from disorder. Nature 459:1063–1064
Tolman JR, Ruan K (2006) NMR residual dipolar couplings as probes of biomolecular dynamics. Chem Rev 106:1720–1736. doi:10.1021/cr040429z
Wöhnert J, Franz KJ, Nitz M et al (2003) Protein alignment by a coexpressed lanthanide-binding tag for the measurement of residual dipolar couplings. J Am Chem Soc 125:13338–13339. doi:10.1021/ja036022d
Author information
Authors and Affiliations
Corresponding author
Additional information
Dominic Barthelmes and Katja Barthelmes have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Barthelmes, D., Barthelmes, K., Schnorr, K. et al. Conformational dynamics and alignment properties of loop lanthanide-binding-tags (LBTs) studied in interleukin-1β. J Biomol NMR 68, 187–194 (2017). https://doi.org/10.1007/s10858-017-0118-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-017-0118-5