Abstract
Riverine environments host diverse microbial communities, exhibiting distinctive assemblies at both microscopic and macroscopic levels. Despite the complexity of microbial life in rivers, the underlying factors that shape the community structure across different compartments remain elusive. Herein, we characterized microbial community composition of biofilm and planktonic (water column) compartments in five naturally saline inland streams and a freshwater stream to examine changes in microbial communities following salinization via sequencing of the microbial 16S rRNA gene. Significant differences in specific conductivity, oxidation–reduction potential, dissolved oxygen, and pH among the sampled streams were measured, as were significant differences in the microbial community composition between the planktonic and biofilm. The bacterial families Bacillaceae, Vicinamibacterceae, and Micrococcaceae were significantly more abundant in the biofilm compartment, while Methylophilaceae, Alcaligenaceae, Spirosomaceae, Burkholderiaceae, and Comamonadaceae were more abundant in the planktonic compartment. In addition, salinity (based on specific conductivity) influenced the microbial community composition in both compartments, with higher sensitivity of the planktonic compartment. Increases in the bacterial families Shewanellaceae, Marinomonadaceae, and Saccharospirillaceae or loss of Anaeromyxobacteraceae could be indicative of increased salinity within inland streams. Our results suggest that monitoring of microbial assemblages of freshwater ecosystems could be used as early warning signs of increased salinization levels.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Global freshwater resources that provide water for human and livestock consumption, industry, and energy are under growing threat from natural and anthropogenic salinization (Jackson et al., 2001; Cañedo-Argüelles et al., 2013; Sujay et al., 2018). Salinization has potentially harmful consequences for ecosystem processes and biological diversity at the macro- and micro-scale. However, the impacts of salinization on aquatic ecological systems remain only partially understood. Typically, salinization changes the ionic contents and pH of natural freshwater systems, and prolonged salinization leads to drastic changes in the ecology and biodiversity of impacted habitats (Sujay et al., 2018). Inland aquatic ecosystems are typically categorized into freshwater or naturally saline systems (Berdugo et al., 2020; Shadrin et al., 2023). Most naturally, saline inland surface ecosystems are lentic systems (lakes, lagoons, etc.), with lotic systems (streams, rivers, etc.) representing a small subcategory (Shadrin et al., 2023).
Categorizing inland lotic surface systems based on salinity (approximated from measured specific conductivity, µS/cm) into either freshwater or naturally saline ecosystems depends on various factors, such as geographic location, underlying geology, mean annual temperature, and precipitation (Cormier et al., 2013). Generally, surface lotic waters across the United States are classified into fresh water (0–1999 µS/cm), slightly saline (2000–5999 µS/cm), moderately saline (6000–19,999 µS/cm), very saline (20,000–69,999 µS/cm), and briny (> 70,000 µS/cm), based on specific conductivity (µS/cm at 25 °C) (Taboga et al., 2018). However, deriving one overall salinity cut-off for lotic surface systems is challenging and not without problems (Cormier et al., 2013). For example, lotic aquatic ecosystems in the eastern USA tend to have, on average, specific conductivity values < 200 µS/cm; in contrast, aquatic ecosystems in the Prairie, Great Basin, and Cold Desert regions in the western USA tend to be higher (> 200 µS/cm) (Griffith and Griffith, 2014). Thus, the cut-off for what qualifies as a freshwater or saline ecosystem in the western USA differs from the east.
Naturally saline, inland, surface ecosystems provide various inherent ecological functions and services despite being different from freshwater ecosystems (Velasco et al., 2006; Paul and Mormile, 2017). These ecosystems are unique in that they maintain exceedingly high salinity characterized by various geological (e.g. evaporite rocks) and climactic (e.g. arid-semiarid) conditions (Last and Ginn, 2005; Millán et al., 2011; Rashed, 2016). One critical value of naturally saline inland surface aquatic ecosystems is that they can provide insights into anticipated changes in freshwater ecosystems following salinization. (Herbert et al., 2015; Kefford et al., 2016; Schuler et al., 2019). Ecosystems such as these provide a glimpse into the kinds and magnitude of changes that can occur in a rapidly warming climate, both in ecosystem functions and microbial diversity, due to alterations in physicochemical parameters (Feeley et al., 2017; Berger et al., 2019; Dudgeon, 2019; Flitcroft et al., 2019; Schuler et al., 2019).
There is a considerable body of work detailing the planktonic microbial community compositions from lentic saline aquatic ecosystems, such as saline lakes (Aanderud et al., 2016; Vavourakis et al., 2016; Han et al., 2017; Naghoni et al., 2017; Hoffman et al., 2018) and coastal wetlands and marshes (Bowen et al., 2009; Chambers et al., 2016; Li et al., 2019; Vera-Gargallo et al., 2019). There is also increasing evidence of distinct compartment-specific microbial communities in non-saline lentic aquatic systems (Eckert et al., 2020; Lambie and Hunter, 2021). These systems are naturally lentic, characterized by some internal stratification, leading to different microbial communities at various depths (Salcher et al., 2011). In contrast, there are far fewer studies of microbial community compositions from lotic saline ecosystems (e.g. streams and rivers) (Lu et al., 2015; Castelán-Sánchez et al., 2019). How well this compartmentalization effect applies to saline lotic systems, such as streams and rivers, remains to be fully understood, although there are data showing differences in compartments within freshwater lotic systems (Zeglin, 2015; Hotaling et al., 2019; Engloner et al., 2023). However, few comparative studies have investigated bacterial community structuring within compartments in lotic freshwater and naturally saline inland aquatic systems. This has potential biodiversity ramifications for freshwater ecosystems facing salinization. Studies like this might also be crucial in detecting and possibly activating mitigation strategies against salinization.
In this study, we sought to characterize in situ microbial communities across the planktonic and biofilm compartments of freshwater and naturally saline inland aquatic ecosystems to get insights into (1) what the microbial community compositions are within the planktonic and biofilm compartments, (2) what microbial taxa might be indicative of these systems undergoing salinization, and (3) where within the compartments of a lotic freshwater system this change might be most notable. To do so, we assessed differences in microbial assemblages between systems that vary naturally in salinity using 16S rRNA amplicon sequencing. These ecosystems are natural experimental gradients (high-salinity, medium-salinity, low-salinity, and freshwater systems) to investigate environmental microbial community composition changes. Overall, we anticipated comparatively higher microbial diversity in the freshwater system because of the constraint on microbial biodiversity that salinity (and the adaptations therein) imposes (Wang et al., 2012; Tang et al., 2021). We also anticipated significant differences in microbial community composition in biofilm and planktonic compartments in both freshwater and saline systems due to the relative stability of biofilm microbial communities compared to planktonic microbial communities (Van Horn, 2011; Portillo et al., 2012; Kamjunke et al., 2015), and we expected the saline systems to be highlighted by the enrichment of different halophilic (salt tolerant) bacterial taxa.
Materials and methods
Site description, sample collection, and processing, and DNA extraction
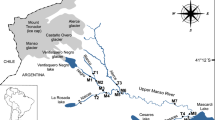
The Powder River Basin is a high-elevation, low-precipitation, and high-temperature shortgrass prairie ecosystem in Wyoming, in the Mountain West region of the USA (Tronstad et al., 2018). The Powder River Basin is in the north-central and eastern parts of Wyoming, with Gillette and Sheridan being the major cities in this basin. It is characterized by Newcastle sandstone geologic formation, and sodium and chloride ions are the significant components of the dissolved salts fraction of the streams within the basin (Taboga et al., 2018). We sampled five natural saline streams in the Powder River Basin. These were Tributary to Murphy Creek (Site 1, S1), Dugout Creek (Site 2, S2), Cloud Creek (Site 3, S3), Dead Horse Creek (Site 4, S4), and the South Fork of the Powder River (Site 5, S5). These saline streams are habitats for the critically endangered and endemic salt-tolerant aquatic beetle species in Wyoming, Hygrotus diversipes (Tronstad, 2015; Tronstad et al., 2018). Because of this, we have opted not to include exact geographic identifiers to preserve the habitats of this endemic species. We also sampled the Laramie River (a freshwater stream in the North Platte River Basin) to compare against the inland saline streams. At two to four locations within each stream, we measured dissolved oxygen concentrations (DO, mg/L), specific conductivity (SPC, µS/cm), pH, and oxidation–reduction potential (ORP, mV), using a Professional Plus multi-probe (YSI Inc. Yellow Springs, OH, USA).
We collected three to four replicate water (planktonic) samples from each stream by submerging sterile plastic containers in the middle of the streams to whatever depth was possible (< 30 cm). These samples were collected to assess the microbial composition of the planktonic community in the water column. We also collected three to four replicates of substrates (various cobbles and gravel) in the middle and along the banks of each stream at varying depths depending on stream conditions using gloves. We transferred substrates into sterile Whirl–Pak bags (Whirl–Pak. Madison, WI, USA) to assess biofilm microbial community composition. Water and substrate samples were stored on ice for transport. In the laboratory, we vacuum-filtered planktonic samples within 24 h of collection by transferring 500 ml into sterile filter containers with 0.22-µm pore Polyether sulfone (PES) membrane filters (Corning Inc. New York, NY, USA). Residue-containing membrane filters were removed with sterile dissecting forceps and placed into a sterile 1.5-ml Eppendorf tube (Fisher Scientific, Pittsburgh, PA, USA). For the biofilm samples, we added 300–500 ml of nanopure water to substrate samples in the Whirl–Pak bags, scrubbed them with sterile brushes for ~ 3 min, and then shook them on a rotating shaker for approximately 10 min. We spent approximately the same time brushing and shaking each bag to standardize the biofilm collection. The resulting solution was transferred into sterile filter bottle systems and vacuum filtered like the water samples. Because the amount of biofilm obtained on the membrane filters varied following filtration across streams, we used approximately one quadrant of each circular membrane filter for DNA extraction for both water and biofilm samples. Planktonic and biofilm membrane filters were stored at – 20 °C until DNA extraction. DNA extraction from water and biofilm membrane samples was carried out using the Qiagen DNeasy PowerWater and DNeasy PowerBiofilm kit, respectively (Qiagen Inc. Germantown, MD, USA), according to the manufacturer’s instructions.
Amplicon library preparation, sequencing, and analyses
DNA extracts from water (planktonic) (n = 22) and biofilm (n = 15) samples were used as templates for amplicon sequencing. Briefly, amplicon libraries of the V4 region of the bacterial 16S rRNA gene were generated using meta-barcoded modified forward (515f) (Parada et al., 2016) and reverse (806r) (Apprill et al., 2015) primers (Walters et al., 2016). We carried out triplicate PCR for all samples using Phusion polymerase and the Phusion High-Fidelity PCR Master Mix with HF Buffer, water, and barcoded primers in a final reaction volume of 20 µl. Reaction conditions consisted of an initial denaturation for 30 s at 98 °C, 30 cycles of 10 s at 98 °C (denaturation), 10 s at 65 °C (annealing), and 8 s at 72 °C (extension), with a final extension phase of 5 min at 72 °C. Using the manufacturer’s protocol, gel-verified amplified products were purified using the Axygen AxyPrep Mag PCR Clean-up kit (Axygen Biosciences, Union City, CA, USA). DNA concentrations were determined using a Qubit 3.0 fluorometer (Invitrogen, Carlsbad, CA, USA). We then pooled equimolar amounts of purified PCR products (12 ng) together. The final library was then sent for sequencing at the University of Minnesota Genomics Center (UMGC, St. Paul. MN, USA) using the Illumina Miseq platform using V2 chemistry (2 × 250 PE). Raw sequence data for all 37 samples are available in the NCBI Sequence Read Archive under BioProject accession number PRJNA598044.
Primary quality checking (filtering and error learning and dereplication) of fastq reads was carried out using DADA2 (Callahan et al., 2016), and primer sequence removal was carried out using cutadapt (Martin, 2011). DADA2 was used for subsequent chimera removal (removeChimeraDenovo) and taxonomy assignments of amplicon sequence variants (ASV) using the SILVA database (version Silva_v138) (Quast et al., 2012). We formatted the final ASV table to include taxonomic information and exported it to a biom format for downstream analyses in QIIME (v.1.9) (Caporaso et al., 2010) and R (R Core Team, 2022). The ASV table was filtered to remove ASVs that were unassigned at the kingdom level, i.e. Bacteria (n = 15), ASVs that were assigned to Eukaryota at the Kingdom level (n = 57), and ASVs unassigned beyond the Bacteria kingdom level (n = 380). We combined replicate biofilm samples collected within the same saline stream with low reads. The final filtered ASV table had a total of 348,214 reads with 11,426 ASVs (0.82% assigned to Archaea and 99.17% assigned to Bacteria) distributed across 37 samples (minimum # of reads = 1037, maximum = 18,899, and mean = 9411 ± 5332 [± Std. deviation]). We rarefied the ASV table to 1,000 reads per sample before all diversity analyses. We assessed alpha- (e.g. richness and evenness) (Simpson, 1949; Shannon C.E, 1957; Chao, 1984) and beta-diversity (Bray–Curtis distance matrix) (Bray and Curtis, 1957) using the rarefied ASV table to determine within-stream and between-stream sample diversity using non-parametric analyses. We used the group significance command in QIIME (v.1.9) to examine the microbial community members (at the family level) potentially driving differences among samples and determined differentially abundant AVS across sample groups (biofilm vs planktonic) and sample categories (salinity level) using non-parametric Kruskal–Wallis test and FDR-corrected p-value. For clarity, groups refer to the planktonic and biofilm samples from all sampled streams and categories refer to the collapsing of sampled stream into freshwater, low-, medium-, and high-salinity stream classifications. We then visualized differences among samples using an NMDS plot.
Statistical analyses
We used the Kruskal–Wallis non-parametric test to examine differences among non-normally distributed variables (alpha level of 0.05), followed by a test for pairwise comparison to see if there were significant stream effects. All analyses were conducted in JMP Pro 14 (SAS Inc. Cary. NC, USA). Following analyses of stream variables, the sampled streams were summarized into low salinity (S5), medium salinity (S2-S4), high salinity (S1), and freshwater (Laramie River, site 6) categories based on measured SPC values (Table 1). Figures were generated using JMP Pro 14 (SAS Inc. Cary. NC, USA).
Results
Stream physicochemical properties
We detected significant differences in specific conductivity (P = 0.01), pH (P = 0.04), dissolved oxygen (P = 0.04), and oxidation–reduction potential (P = 0.01) among the six sampled streams, with higher mean values in all the five saline streams relative to the freshwater stream (Table 1A). The mean specific conductivity in all five saline streams was (14,748.9 ± 8228.5, mean ± std) relative to the freshwater stream (663 ± 498). Similarly, mean dissolved oxygen (7.87 ± 2.40), oxidation–reduction potential (46.2 ± 23.4), and pH (8.34 ± 0.36) were significantly higher in all five saline streams relative to the freshwater stream (Table 1A). Furthermore, we detected significant differences in specific conductivity (P = 0.005), dissolved oxygen (P = 0.05), pH (P = 0.02), and oxidation–reduction potential (P = 0.005) among low salinity, medium salinity, high salinity, and freshwater system categories (Table 1B). The high salinity stream category had significantly higher mean specific conductivity, dissolved oxygen, and oxidation–reduction potential relative to the low- and medium-salinity and freshwater streams (Table 1B) but had a significantly lower pH than the other stream categories.
Microbial diversity and community structure
There was an overall significant difference in microbial diversity between compartments (biofilm and planktonic) across all sampled streams (n = 2 groups; Simpson’s index, P < 0.0001; Shannon diversity, P = 0.0002). Biofilm samples (from both saline and freshwater streams) were significantly richer in microbial species relative to associated planktonic (water) samples (Fig. 1A). Microbial diversity in sampled streams categories tended to be comparable across both biofilm and planktonic compartments, except a lower Shannon diversity for the high-salinity biofilm compartment compared to the low- and medium-salinity biofilm compartments (Fig. 1B). Furthermore, there were no significant differences in microbial alpha-diversity among the four stream categories (freshwater, low salinity, medium salinity, high salinity) based on two of the alpha-diversity indices used (n = 4 stream types; Simpson’s index, P = 0.12; Shannon diversity, P = 0.10) (Fig. 1B). However, Simpson’s and Shannon diversity indices were significantly different across individual streams and their compartments (n = 12 groups; Simpson’s index, P = 0.002; Shannon diversity, P = 0.01) (Fig. 1C).
A Differences in microbial species diversity (Simpson’s index and Shannon diversity) across biofilm (blue) and planktonic (green) stream compartments in all sampled streams. B Differences in microbial diversity (Simpson’s index and Shannon diversity) in biofilm and plankton compartments of the sampled stream categories (Freshwater, low-, medium-, high-salinity streams) based on specific conductivity. C Differences in microbial diversity (Simpson’s index and Shannon diversity) in biofilm and plankton compartments of stream types (each of the five saline streams and the freshwater stream). Different letters indicate significant differences at P < 0.05. Box plot represents the median and the interquartile range
For beta diversity, first, we examined the Bray–Curtis distance matrix for both compartments for variances and uncovered significant differences among compartments (Betadisper, P < 0.001; PERMDISP, P < 0.001), with higher dispersion in the biofilm compared to planktonic compartment (Fig. 2A). Examining the influence of stream variables on community composition revealed a significant correlation between specific conductivity and NMDS axis 3 (Adjusted R2 = 0.52, P < 0.001), indicating a significant impact of specific conductivity on microbial community composition. Overall, NMDS axis 1 separated samples according to compartments (PERMANOVA test-statistic = 18.76, P < 0.001) (Fig. 2A), and NMDS axis 3 further separated samples according to specific conductivity. The influence of salinity (specific conductivity) was also confirmed with a PERMANOVA analysis based on streams categories (freshwater, low salinity, medium salinity, and high salinity) (PERMANOVA test-statistic = 6.68, P < 0.001) (Fig. 2B). However, there was no interaction between compartment and stream category (Adonis, R2 = 0.11, P = 0.11), indicating salinity affected both compartments similarly. Given the strong effect of compartment on microbial community composition, an investigation of the factors driving microbial community composition within each compartment was done. Within the biofilm compartment, stream categorization based on specific conductivity (fresh, low, medium, and high salinity) did not have a significant effect on microbial community composition (Adonis, R2 = 0.27, P = 0.08) (Fig. 2B). In contrast, in the planktonic compartment stream categorization did have a significant effect on microbial composition (Adonis, R2 = 0.28, P = 0.03) (Fig. 2B), with the medium-salinity stream differing from the high-salinity stream samples (P = 0.03) and marginally significantly from the low-salinity (P = 0.07) and freshwater stream (P = 0.08) samples (Table S2). In addition, regression analysis between the environmental variable specific conductivity and NMDS axis 3 confirmed the stronger effect of salinity within the planktonic (R2 = 0.93, P < 0.0001) compared to the biofilm compartment (R2 = 0.26, P = 0.05).
Beta-diversity summaries from sampled sites. NMDS plots showing within-stream differences in bacterial community composition of the biofilm (blue) and planktonic (green) stream compartments across A all sampled streams and B sampled streams categorized according to measured SPC values. Different markers indicate different streams (sites) in A and stream categories in B. Stress for plots A and B is 0.06. Ellipses represent a 95% confidence interval of correctly assigning samples into groups
Differences in bacterial community composition between the biofilm and planktonic compartments were underscored by 52 differentially abundant bacterial families (Table S1A). In the biofilm compartment, Unassigned Bacilli, Vicinamibacteraceae, Unassigned Vicinamibacteraceae, WD2101_Soil_group, Pirellulaceae, Pseudonocardiaceae, Nocardioidaceae, Halomonadaceae, Micrococcaceae, Chthoniobacteraceae, Crocinitomicaceae, Caulobacteraceae, Blastocatellaceae, Bacillaceae, AKYG1722, Anaerolineaceae, Solirubrobacteraceae, Solirubrobacterales (Family 67–14), and Anaerolineae (Family A4b) were in higher relative abundance compared to the planktonic compartment (Fig. 3A, Table S1A). In contrast, Methylophilaceae, Alcaligenaceae, Spirosomaceae, Burkholderiaceae, Sporichthyaceae, Sphingobacteriaceae, Microbacteriaceae, Comamonadaceae, Crocinitomicaceae, Flavobacteraceae, and Microbacteriaceae were in higher relative abundance in the planktonic compartment compared to the biofilm compartment (Fig. 3A, Table S1A).
Most of these same bacterial families also influenced differences in community composition across stream categories (freshwater, low, medium, high salinity) in biofilm and planktonic compartments, with some additional taxa emerging (Fig. 3B, Table S1B). The families Shewanellaceae, Marinomonadaceae, and Saccharospirillaceae were indicative of increased salinity with higher relative abundance in the high-salinity biofilm and planktonic samples, and Halomonadaceae was in higher relative abundance in medium- and high-salinity biofilm samples compared to the low-salinity and freshwater samples (Fig. 3B, Table S1B). In contrast, Anaeromyxobacteraceae was indicative of low salinity with higher relative abundance in freshwater biofilm samples compared to the saline samples (Fig. 3B, Table S1B). In addition, some families were indicative of low- to medium-salinity levels: Alcaligenaceae was present in significantly higher amounts in the low- and medium-salinity planktonic samples compared to the other planktonic samples, and AKYG1722 had higher relative abundance in low-salinity and medium-salinity biofilm samples compared to the other biofilm samples (Fig. 3B, Table S1B).
Discussion
The potential threat of salinization to freshwater ecosystem functions and services is a major global ecological issue; however, the anticipated impacts of increasing salinization on microbial assemblages in freshwater ecosystems are varied and unclear. In this study, we sought to capture potential differences in microbial assemblages in the biofilm and planktonic compartments of natural freshwater lotic systems and saline inland aquatic stream ecosystems to provide insight into what these changes might be because of increasing salinization.
The microbial community composition in lotic saline or freshwater systems can be impacted by the nutrient status of the system, the presence and type of organic matter in the system, and available surfaces for bacterial attachment. This creates spatially different regions within streams for microbes to assemble and colonize (Zeglin, 2015). It is known that specific bacterial taxa are generally more enriched in biofilm compartments within freshwater systems than in the bacterioplankton/water column compartment due to the relative stability of biofilm microbial communities compared to planktonic microbial communities (Van Horn, 2011; Portillo et al., 2012; Kamjunke et al., 2015; Zeglin, 2015). In this study, we confirmed that naturally saline inland streams behave similarly showing differences in planktonic (water) and biofilm microbial communities of the sampled streams underscored by differentially abundant bacterial families (per aim one of this study). Biofilm samples in this study were differentially enriched in ~ 20 families, with abundant ones in the phyla Actinobacteriota (family Micrococcaceae, Solirubrobacteraceae, Nocardioidaceae), Acidobacteroidota (family Unassigned Vicinamibacterales, Vicinamibacteraceae), Bacteroidetes/Bacteroidota (family, Microscillaceae, Cryomorphaceae, Unassigned Bacteroidota), and members of the superphylum PVC (family Chthoniobacteraceae, Pirellulaceae, WD2101_soil_group) (Fig. 3A, Table S1A). These bacterial taxa have been previously reported as abundant in biofilm compartments relative to planktonic samples (Bengtsson and Øvreås, 2010; Martiny et al., 2015; Zeglin, 2015; Hotaling et al., 2019; Ren et al., 2020). In contrast, the planktonic compartments were enriched in ~ 9 families that included previously reported members of the Actinobacteria (Microbacteriaceae and Sporichthyaceae), Bacteroidota (Spirosomaceae, Sphingobacteriaceae, Flavobacteriaceae), and Proteobacteria (Burkholderiaceae, Oxalobacteriaceae, and Rhodobacteriaceae) (Portillo et al., 2012; Zeglin, 2015; Ayayee et al., 2018). The underlying cause for this distinct clustering of biofilm and planktonic samples may be attributed to the relative stability and resource utilization ability of biofilm microbial communities compared to planktonic microbial communities in freshwater systems (Van Horn, 2011; Portillo et al., 2012; Kamjunke et al., 2015). Our data suggest that this may also be the case in naturally saline inland aquatic systems based on the higher alpha-diversity observed in the biofilm compared to the planktonic compartment, with higher diversity providing greater resource utilization ability.
We also determined significant differences in microbial assemblages between the saline and freshwater systems sampled, for the planktonic compartment (Fig. 2B). Given this study’s large range of specific conductivity from ~ 9000 to 25,000 µS/cm in the five sampled inland saline streams relative to ~ 660 µS/cm in the freshwater stream, effects on microbial communities were expected. Our study confirms the well-documented impacts of salinity on habitat quality and ecology of aquatic life (Kefford et al., 2016; Sujay et al., 2018; Berger et al., 2019). Interestingly, although salinity affected the bacterial community composition, we did not see any effects of salinity on bacterial alpha-diversity (Fig. 1B). It is anticipated that following the salinization of freshwater ecosystems, bacterial taxa typically associated with saline ecosystems will become more emergent, indicative of changed ecosystems, as well as the constraints imposed on microbial diversity (and the adaptations therein) because of increasing salinization (Wang et al., 2012; Tang et al., 2021). Previously reported halophilic (salt tolerant) taxa from various sources, including naturally saline inland streams, such as unassigned Bacilli, Comamonadaceae, Methylophilaceae, and Alcaligenaceae (Vavourakis et al., 2016; Han et al., 2017; Jacob et al., 2017; Naghoni et al., 2017; Cabello-Yeves and Rodriguez-Valera, 2019; Salcher et al., 2019) were recovered in higher abundances from both planktonic and biofilm compartments of saline streams relative to the freshwater streams in this study. This result may reflect the high-salt concentrations in the inland saline streams sampled in this study and the selection of salt-tolerant microbial families within them. Other less-known salt-tolerant bacterial taxa, namely Sporichthyaceae (Vavourakis et al., 2016; Yilmaz et al., 2016) and Spirosomaceae (Rojas et al., 2021), were also statistically more abundant in both biofilm and planktonic compartments of our saline microbial community data. Conversely, several taxa previously reported from freshwater systems, such as Anaeromyxobacteraceae and T34 (Newton et al., 2011; Schultz Jr. et al., 2013; Carney et al., 2015, 2016; Sun et al., 2017; Tessler et al., 2017; Akins et al., 2018; Ayayee et al., 2018; Chiang et al., 2018; Hotaling et al., 2019) were also unearthed in statistically higher abundances in both biofilm and planktonic compartments of the freshwater stream in this study. This further highlights the selection of non-halophilic bacterial families in freshwater systems (per aim two of this study).
Regarding where within a lotic freshwater system change in microbial community composition following salinization might be most notable in a freshwater system, our data suggest that the planktonic compartment might be an appropriate place, since we observed the strongest effect of salinity on microbial community composition in this compartment. We observed significant differential abundances of previously reported salt-tolerant bacterial taxa such as Micrococcaceae and Sphingobacteriaceae in the planktonic compartment of sampled freshwater stream, as well as saline stream planktonic compartments in this study. Interestingly, we also observed a high abundance of halophilic bacterial taxa, such as AKYG1722, in the biofilm compartment of the sampled freshwater stream (Table S1B). Taken together, both the presence and abundance of such taxa in the biofilms of freshwater systems may indicate a freshwater system undergoing salinization (although the planktonic compartment nonetheless seem to be comparatively more impacted by salinity). This may vary from system to system, but it provides a way to gauge and evaluate a system undergoing salinity in response to myriad factors (as per aim three of this study). Given that planktonic communities are less stable and experience more turnover than biofilm communities, the biofilm compartments may represent more severe and permanent changes to microbial communities in response to salinity, given the way they are structured and impacted by processes within the lotic system (Portillo et al., 2012; Kamjunke et al., 2015; Hotaling et al., 2019) and their relative higher resilience. Thus, detecting salt-tolerant bacterial taxa in this compartment might indicate more than just a sporadic occurrence and could underscore a changing system.
Conclusion
This study uncovered significant differences in the bacterial community composition of planktonic (water column) and biofilm compartments of five naturally saline streams and one freshwater stream. We detected significant differences in community composition based on salinity level, with the largest salinity effect in the planktonic compartment. This confirms our expectation since these sampled streams varied significantly in underlining physicochemical variables, such as specific conductivity. However, the salinity effects were not reflected in bacterial alpha diversity metrics. The data generated in this study provides a framework for further studies investigating predicted changes in freshwater microbial community compositions because of increasing salinization.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
References
Aanderud, Z. T., J. C. Vert, J. T. Lennon, T. W. Magnusson, D. P. Breakwell & A. R. Harker, 2016. Bacterial Dormanncy Is More Prevalent in Freshwater tha Hypersaline Lakes. Front. Microbiol. 7: 853. https://doi.org/10.3389/fmicb.2016.00853.
Apprill, A., McNally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton . Aquat. Microb. Ecol. 75, 129–137. Available at: https://www.int-res.com/abstracts/ame/v75/n2/p129-137/.
Ayayee, P. A., C. R. Cosgrove, A. Beckwith, A. A. Roberto & L. G. Leff, 2018. Gut bacterial assemblages of freshwater macroinvertebrate functional feeding groups. Hydrobiologia 822: 157–172. https://doi.org/10.1007/s10750-018-3671-3.
Bengtsson, M. M. & L. Øvreås, 2010. Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol. 10: 261. https://doi.org/10.1186/1471-2180-10-261.
Berdugo, M., M. Delgado-Baquerizo, S. Soliveres, R. Hernández-Clemente, Y. Zhao, J. J. Gaitán, et al., 2020. Global ecosystem thresholds driven by aridity. Science. 367: 787–790. https://doi.org/10.1126/science.aay5958.
Berger, E., Frör, O., and Schäfer, R. B. (2019). Salinity impacts on river ecosystem processes: a critical mini-review. Philos. Trans. R. Soc. B Biol. Sci. 374, 20180010. doi:https://doi.org/10.1098/rstb.2018.0010.
Bowen, J. L., B. C. Crump, L. A. Deegan & J. E. Hobbie, 2009. Salt marsh sediment bacteria: their distribution and response to external nutrient inputs. ISME J. 3: 924–934. https://doi.org/10.1038/ismej.2009.44.
Bray, J. R. & J. T. Curtis, 1957. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 27: 325–349. https://doi.org/10.2307/1942268.
Cabello-Yeves, P. J. & F. Rodriguez-Valera, 2019. Marine-freshwater prokaryotic transitions require extensive changes in the predicted proteome. Microbiome 7: 117. https://doi.org/10.1186/s40168-019-0731-5.
Callahan, B. J., C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer, et al., 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13: 581–583. https://doi.org/10.1038/nmeth.3869.
Cañedo-Argüelles, M., B. J. Kefford, C. Piscart, N. Prat, R. B. Schäfer & C.-J. Schulz, 2013. Salinisation of rivers: an urgent ecological issue. Environ. Pollut. 173: 157–167. https://doi.org/10.1016/j.envpol.2012.10.011.
Caporaso, J. G., J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, et al., 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7: 335–336. https://doi.org/10.1038/nmeth.f.303.
Castelán-Sánchez, H. G., P. Elorrieta, P. Romoacca, A. Liñan-Torres, J. L. Sierra, I. Vera, et al., 2019. Intermediate-Salinity Systems at High Altitudes in the Peruvian Andes Unveil a High Diversity and Abundance of Bacteria and Viruses. Genes (basel). 10: 891. https://doi.org/10.3390/genes10110891.
Chambers, L. G., R. Guevara, J. N. Boyer, T. G. Troxler & S. E. Davis, 2016. Effects of Salinity and Inundation on Microbial Community Structure and Function in a Mangrove Peat Soil. Wetlands 36: 361–371. https://doi.org/10.1007/s13157-016-0745-8.
Chao, A., 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11: 265–270.
Cormier, S. M., G. W. Suter II. & L. Zheng, 2013. Derivation of a benchmark for freshwater ionic strength. Environ. Toxicol. Chem. 32: 263–271. https://doi.org/10.1002/etc.2064.
Dudgeon, D., 2019. Multiple threats imperil freshwater biodiversity in the Anthropocene. Curr. Biol. 29: R960–R967. https://doi.org/10.1016/j.cub.2019.08.002.
Eckert, E. M., S. Amalfitano, A. Di Cesare, C. Manzari, G. Corno & D. Fontaneto, 2020. Different substrates within a lake harbour connected but specialised microbial communities. Hydrobiologia 847: 1689–1704. https://doi.org/10.1007/s10750-019-04068-1.
Engloner, A. I., M. Vargha, P. Kós & A. K. Borsodi, 2023. Planktonic and epilithic prokaryota community compositions in a large temperate river reflect climate change related seasonal shifts. PLoS One 18: e0292057. https://doi.org/10.1371/journal.pone.0292057.
Feeley, H., M. Bruen, C. Bullock, M. Christie, F. Kelly, K. Remoundou, et al., 2017. Freshwater Ecosystem Services - an Introduction for Stakeholders. https://doi.org/10.13140/RG.2.2.28622.25928.
Flitcroft, R., M. S. Cooperman, I. J. Harrison, D. Juffe-Bignoli & P. J. Boon, 2019. Theory and practice to conserve freshwater biodiversity in the Anthropocene. Aquat. Conserv. Mar. Freshw. Ecosyst. 29: 1013–1021. https://doi.org/10.1002/aqc.3187.
Griffith, M. B. & M. B. Griffith, 2014. Natural Variation and Current Reference for Specific Conductivity and Major Ions in Wadeable Streams of the Conterminous USA. Freshw. Sci. 33: 1–17. https://doi.org/10.1086/674704.
Han, R., X. Zhang, J. Liu, Q. Long, L. Chen, D. Liu, et al., 2017. Microbial community structure and diversity within hypersaline Keke Salt Lake environments. Can. J. Microbiol. 63: 895–908. https://doi.org/10.1139/cjm-2016-0773.
Herbert, E. R., P. Boon, A. J. Burgin, S. C. Neubauer, R. B. Franklin, M. Ardon, et al., 2015. A global perspective on wetland salinization: Ecological consequences of a growing threat to freshwater wetlands. Ecosphere 6: 1. https://doi.org/10.1890/ES14-00534.1.
Hoffman, S. K., K. W. Seitz, J. C. Havird, D. A. Weese & S. R. Santos, 2018. Diversity and the environmental drivers of spatial variation in Bacteria and micro-Eukarya communities from the Hawaiian anchialine ecosystem. Hydrobiologia 806: 265–282. https://doi.org/10.1007/s10750-017-3365-2.
Hotaling, S., M. E. Foley, L. H. Zeglin, D. S. Finn, L. M. Tronstad, J. J. Giersch, et al., 2019. Microbial assemblages reflect environmental heterogeneity in alpine streams. Glob. Chang. Biol. 25: 2576–2590. https://doi.org/10.1111/gcb.14683.
Jackson, R. B., S. R. Carpenter, C. N. Dahm, D. M. McKnight, R. J. Naiman, S. L. Postel, et al., 2001. Water in a Changing World. Ecol. Appl. 11: 1027–1045. https://doi.org/10.2307/3061010.
Jacob, J. H., E. I. Hussein, M. A. K. Shakhatreh & C. T. Cornelison, 2017. Microbial community analysis of the hypersaline water of the Dead Sea using high-throughput amplicon sequencing. Microbiologyopen 6: e00500. https://doi.org/10.1002/mbo3.500.
Kamjunke, N., P. Herzsprung & T. R. Neu, 2015. Quality of dissolved organic matter affects planktonic but not biofilm bacterial production in streams. Sci. Total Environ. 506–507: 353–360. https://doi.org/10.1016/j.scitotenv.2014.11.043.
Kefford, B. J., D. Buchwalter, M. Cañedo-Argüelles, J. Davis, R. P. Duncan, A. Hoffmann, et al., 2016. Salinized rivers: degraded systems or new habitats for salt-tolerant faunas? Biol. Lett. 12: 20151072. https://doi.org/10.1098/rsbl.2015.1072.
Lambie, S. M. & D. W. F. Hunter, 2021. Microbial composition in different physical compartments of six constructed wetland systems in New Zealand. Ecol. Eng. 166: 106238. https://doi.org/10.1016/j.ecoleng.2021.106238.
Last, W. M. & F. M. Ginn, 2005. Saline systems of the Great Plains of western Canada: an overview of the limnogeology and paleolimnology. Saline Systems 1: 10. https://doi.org/10.1186/1746-1448-1-10.
Li, W., X. Lv, J. Ruan, M. Yu, Y.-B. Song, J. Yu, et al., 2019. Variations in Soil Bacterial Composition and Diversity in Newly Formed Coastal Wetlands. Front. Microbiol. 9: 3256. https://doi.org/10.3389/fmicb.2018.03256.
Lu, Z., L. Li, L. Liu, X. Zhang, Y. Yang, J. Li, et al., 2015. Annual periodicity in planktonic bacterial and archaeal community composition of eutrophic Lake Taihu. Sci. Rep. 5: 15488. https://doi.org/10.1038/srep15488.
Martin, M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet.journal 17: 10. https://doi.org/10.14806/ej.17.1.200.
Martiny, J. B. H., S. E. Jones, J. T. Lennon & A. C. Martiny, 2015. Microbiomes in light of traits: A phylogenetic perspective. Science. https://doi.org/10.1126/science.aac9323.
Millán, A., J. Velasco, C. Gutiérrez-Cánovas, P. Arribas, F. Picazo, D. Sánchez-Fernández, et al., 2011. Mediterranean saline streams in southeast Spain: What do we know? J. Arid Environ. 75: 1352–1359. https://doi.org/10.1016/j.jaridenv.2010.12.010.
Naghoni, A., G. Emtiazi, M. A. Amoozegar, M. S. Cretoiu, L. J. Stal, Z. Etemadifar, et al., 2017. Microbial diversity in the hypersaline Lake Meyghan. Iran. Sci. Rep. 7: 11522. https://doi.org/10.1038/s41598-017-11585-3.
Parada, A. E., D. M. Needham & J. A. Fuhrman, 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18: 1403–1414. https://doi.org/10.1111/1462-2920.13023.
Paul, V. G. & M. R. Mormile, 2017. A case for the protection of saline and hypersaline environments: a microbiological perspective. FEMS Microbiol. Ecol. https://doi.org/10.1093/femsec/fix091.
Portillo, C. M., P. Anderson & S., and Fierer, N., 2012. Temporal variability in the diversity and composition of stream bacterioplankton communities. Environ. Microbiol. 14: 2417–2428. https://doi.org/10.1111/j.1462-2920.2012.02785.x.
Quast, C., E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer, P. Yarza, et al., 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41: D590–D596. https://doi.org/10.1093/nar/gks1219.
R Core Team (2022). R: A language and environment for statistical computing. R Found. Stat. Comput. Vienna, Austria. URL http//www.R-project.org/.
Rashed, R. D. W. E.-M. N. (2016). “Lake La Salada de Chiprana (NE Spain), an Example of an Athalassic Salt Lake in a Cultural Landscape,” in (Rijeka: IntechOpen), Ch. 3. doi:https://doi.org/10.5772/64443.
Ren, Z., D. Niu, P. Ma, Y. Wang, Z. Wang, H. Fu, et al., 2020. Bacterial Communities in Stream Biofilms in a Degrading Grassland Watershed on the Qinghai-Tibet Plateau. Front. Microbiol. 11: 1021. https://doi.org/10.3389/fmicb.2020.01021.
Rojas, J., A. M. Binoy, C. Suarez, S. Ratering & S. Schnell, 2021. Spirosoma endbachense sp. nov., isolated from a natural salt meadow. Int. J. Syst. Evol. Microbiol. 71: 1–7. https://doi.org/10.1099/ijsem.0.004601.
Salcher, M. M., J. Pernthaler, N. Frater & T. Posch, 2011. Vertical and longitudinal distribution patterns of different bacterioplankton populations in a canyon-shaped, deep prealpine lake. Limnol. Oceanogr. 56: 2027–2039. https://doi.org/10.4319/lo.2011.56.6.2027.
Salcher, M. M., D. Schaefle, M. Kaspar, S. M. Neuenschwander & R. Ghai, 2019. Evolution in action: habitat transition from sediment to the pelagial leads to genome streamlining in Methylophilaceae. ISME J. 13: 2764–2777. https://doi.org/10.1038/s41396-019-0471-3.
Schuler, M. S., Cañedo-Argüelles, M., Hintz, W. D., Dyack, B., Birk, S., and Relyea, R. A. (2019). Regulations are needed to protect freshwater ecosystems from salinization. Philos. Trans. R. Soc. B Biol. Sci. 374, 2018009. doi:https://doi.org/10.1098/rstb.2018.0019.
Shadrin, N., E. Anufriieva & G. Gajardo, 2023. Ecosystems of Inland Saline Waters in the World of Change. Water. https://doi.org/10.3390/w15010052.
Shannon, C. E., 1957. A Mathematical Theory of Communication. Bell Syst. Tech. J. 27: 379–423. https://doi.org/10.1016/S0006-3495(96)79210-X.
Simpson, E. H., 1949. Measurement of Diversity. 163: 688–688.
Sujay., K., E., L. G., L., P. M., M., U. R., Shahan, H., Julia, G., et al., 2018. Freshwater salinization syndrome on a continental scale. Proc. Natl. Acad. Sci. 115: E574–E583. https://doi.org/10.1073/pnas.1711234115.
Taboga, K., Stafford, J., and Rodger, J. (2018). Groundwater Salinity in the Powder River Basin, Wyoming. Wyoming State Geol. Surv. Open File Rep. 5, 0–23.
Tang, X., Xie, G., Shao, K., Tian, W., Gao, G., and Qin, B. (2021). Aquatic Bacterial Diversity, Community Composition and Assembly in the Semi-Arid Inner Mongolia Plateau: Combined Effects of Salinity and Nutrient Levels. Microorganisms . doi:https://doi.org/10.3390/microorganisms9020208.
Tronstad, L. M., K. M. Brown & M. D. Andersen, 2018. Using Species Distribution Models to Guide Field Surveys for an Apparently Rare Aquatic Beetle. J. Fish Wildl. Manag. 9: 330–339. https://doi.org/10.3996/112016-JFWM-085.
Van Horn, D., 2011. Response of heterotrophic stream biofilm communities to a gradient of resources. Aquat. Microb. Ecol. 64: 149–161. https://doi.org/10.3354/ame01515.
Vavourakis, C. D., R. Ghai, F. Rodriguez-Valera, D. Y. Sorokin, S. G. Tringe, P. Hugenholtz, et al., 2016. Metagenomic Insights into the Uncultured Diversity and Physiology of Microbes in Four Hypersaline Soda Lake Brines. Front. Microbiol. 7: 211. https://doi.org/10.3389/fmicb.2016.00211.
Velasco, J., A. Millán, J. Hernández, C. Gutiérrez, P. Abellán, D. Sánchez, et al., 2006. Response of biotic communities to salinity changes in a Mediterranean hypersaline stream. Saline Systems 2: 12. https://doi.org/10.1186/1746-1448-2-12.
Vera-Gargallo, B., T. R. Chowdhury, J. Brown, S. J. Fansler, A. Durán-Viseras, C. Sánchez-Porro, et al., 2019. Spatial distribution of prokaryotic communities in hypersaline soils. Sci. Rep. 9: 1769. https://doi.org/10.1038/s41598-018-38339-z.
Walters, W., Hyde, E. R., Berg-Lyons, D., Ackermann, G., Humphrey, G., Parada, A., et al. (2016). Improved Bacterial 16S rRNA Gene (V4 and V4–5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems doi:https://doi.org/10.1128/mSystems.00009-15.
Wang, Y., H.-F. Sheng, Y. He, J.-Y. Wu, Y.-X. Jiang, N.F.-Y. Tam, et al., 2012. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl. Environ. Microbiol. 78: 8264–8271. https://doi.org/10.1128/AEM.01821-12.
Yilmaz, P., P. Yarza, J. Z. Rapp & F. O. Glöckner, 2016. Expanding the World of Marine Bacterial and Archaeal Clades. Front. Microbiol. 6: 1524. https://doi.org/10.3389/fmicb.2015.01524.
Zeglin, L. H., 2015. Stream microbial diversity in response to environmental changes: Review and synthesis of existing research. Front. Microbiol. 6: 454. https://doi.org/10.3389/fmicb.2015.00454.
Acknowledgements
The Microbial Ecology Collaborative provided funding for this study at the University of Wyoming with funding from NSF award #EPS‐1655726.
Funding
This article was funded by NSF award #EPS‐1655726, EPS‐1655726, Linda van Diepen.
Author information
Authors and Affiliations
Contributions
PAA, LVD, and LT conceived and designed the study. LT provided information and data on the location of saline streams. PAA and LT collected samples from streams. PAA processed and prepared samples for sequence analyses. PAA, GC, and LVD analyzed the data. PAA wrote the manuscript with input from co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing or financial interests.
Additional information
Handling editor: Stefano Amalfitano
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ayayee, P.A., Custer, G.F., Tronstad, L.M. et al. Unveiling salinity-driven shifts in microbial community composition across compartments of naturally saline inland streams. Hydrobiologia 851, 2627–2639 (2024). https://doi.org/10.1007/s10750-024-05479-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-024-05479-5