Abstract
Freshwater zooplankter Brachionus plicatilis is able to inhabit different habitats and locally adapt to their environmental conditions. It also shows a high degree of population structuring in small geographical regions. Here we try to shed light on the evolution of reproductive isolation in populations of B. plicatilis with presumptive gene flow among locally adapted populations. We have conducted laboratory experiments on admixed pairwise populations that differ in predictability of the water regime. We have assessed the potential for within-population reproductive preferences as a deviation of genotypes from Hardy–Weinberg equilibrium in diapausing eggs, a product of sexual reproduction. We expected heterozygote deficit to increase with environmental distance. We have found signs for incipient reproductive isolation in one third of our admixed populations, however no correlation with environmental distance was found, nor with genetic or geographic predictor variables. The overall inbreeding coefficient showed a tendency for within-population crosses preferences to decrease over time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding the factors and mechanisms that underline population diversification and speciation remains to this day one of the challenges in current biology, due to the complexity of the processes involved. The evolution of reproductive isolation is crucial for the maintenance of population differentiation and, in some cases, subsequent species formation (Mayr, 1963; Coyne & Orr, 2004; Westram et al., 2022). Reproductive isolation arises as a result of different processes or events. The main factors involved in the development of reproductive isolation are sufficiently long periods in allopatry, adaptation to local environments with subsequent selection for reproductive isolation, and historical processes related to range expansion and colonization (Tregenza, 2002; Coyne & Orr, 2004).
In populations separated by large geographical distances, where low or no exchange of individuals occurs, reproductive isolation is expected to arise randomly, as a part of overall population differentiation and processes such as genetic drift or genetic hitchhiking (Mayr, 1963; Coyne & Orr, 2004; Feder et al., 2013). The situation is different without geographical isolation, where exchange of individuals occurs, and subsequent gene flow could continuously disrupt diverging evolution among populations. Here, selection for reproductive isolation can follow incipient local adaptation in order to avoid interpopulation breeding if outbred offspring show lower fitness due to disruption of locally adapted genomes (Butlin, 1987; Coyne & Orr, 2004). Reproductive isolation is in this case likely to occur in earlier phases of species formation (Coyne & Orr, 1989; Yukilevich, 2012; Nosil, 2013). Despite the abundance of theoretical work studying evolution of reproductive isolation, relatively little experimental studies are investigating this topic in its breadth (but see Chin et al., 2019).
Environmental gradients in geographical proximity are convenient to study evolution of reproductive isolation arising due to local adaptation. In the Iberian Peninsula, the facultative sexual zooplankter Brachionus plicatilis (Müller, 1786) inhabits brackish ponds ranging from ephemeral puddles to permanent lakes, embracing wide scales of salinity, temperature, food quality, etc. The Iberian populations of B. plicatilis have been found possessing genetic differences in their traits for timing for sexual reproduction in order to match local environmental unpredictability, as assessed by non-regular fluctuations in the length of annual periods a pond is flooded (Franch-Gras et al., 2017a, b). Populations of B. plicatilis consist of clones of asexual females reproducing by parthenogenetic propagation until sexual reproduction is induced in response to population density (Carmona et al., 1993; Stelzer & Snell, 2003; Gilbert, 2007). As a result of sexual reproduction, diapausing (resting) eggs are produced, and they sink to the sediment, forming a reservoir that allows rotifers to survive adverse periods of the annual cycle. The timing of sexual reproduction (i.e. switching to sexual reproduction earlier or later) is a relevant trait for local adaptation, as switching too early or too late has respectively costs in terms of decreased clonal proliferation or not producing stages able to survive adverse periods. This within-species ecological divergence occurs in a geographical area limited to a few tens of kilometres. Despite the advantages of rotifers as model organisms in micro-evolutionary studies (Declerck & Papakostas, 2016), and contrary to numerous studies on between-species reproductive isolation (Gomez & Serra, 1995; Suatoni et al., 2006; Schröder & Walsh, 2007; Kordbacheh et al., 2019, 2023; Zhang & Declerck, 2022a, b) to date, little attention has been devoted to study the diversification and reproductive isolation within a single species. In order to address the processes involved in the evolution of the reproductive isolation, we advocate for a population approach, with studies including within –and among– population genetic variation which are scant (but see, e.g., Jezkova et al., 2022a, b). A population approach accounts for the genetic variability harboured in local populations, which is important when dealing with quantitative and polygenic traits, as presumably involved in partial reproductive isolation.
With the aid of mating choice experiments, Jezkova et al. (2022a) reported mating behaviour that promotes reproductive isolation between Iberian populations of B. plicatilis, a trend stronger in populations with a higher degree of adaptive divergence to unpredictability. However, this study tested for isolation (1) using high male and female densities (2) in absence of mating competition (i.e., females of one single clone are assayed with males of one single clone), (3) simulating the synchronic timing of sex of the assayed clones. Here, we approach a more natural setup by using laboratory cultures that mix genotypes from two different populations at lower population densities and, unlike in Jezkova et al. (2022a), who studied copulation rates, we look for evidence of reproductive isolation assessed by genetic marker inspection in the diapausing eggs produced. Focussing on this stage results in a composite measure that includes a variety of barriers like assortative mating, fertilization success or even early abortion of embryos. We classified these eggs as produced by intrademic and interdemic crosses. Mixed pairs of natural populations covered a range of ecological divergence. Our hypotheses are that genotypes are prone to intrademic reproduction, and that this tendency is stronger when populations are more distant in the predictability of their environment.
Materials and methods
Study area
The nine studied populations of B. plicatilis inhabit in a small area of Eastern Spain (Table 1). They differ in the length and predictability of the flooding season of the habitats (Franch-Gras et al., 2017a) and are locally adapted to these conditions by having different propensities to initiate sexual reproduction (Franch-Gras et al., 2017b). Additionally, they have been characterized in terms of genetic differentiation with neutral (i.e., microsatellites) and non-neutral markers (i.e., the gene coding for the mate recognition protein, mmr-b, which is responsible for the female-male encounter; Jezkova et al., 2022b).
Culture conditions and clone isolation
Rotifers were cultured at 12 g l−1 artificial sea water (Instant Ocean® Sea Salt, Aquarium Systems), maintained at 20°C with moderate aeration, constant illumination of approx. 35 µmol quanta m−2 s−1. They were fed ad libitum once a week with the microalgae Tetraselmis suecica at 1·106 cells ml−1 which was cultured under the same laboratory conditions as the rotifers.
In order to establish single-clone cultures, diapausing eggs were extracted from the pond sediments using a sugar flotation technique (Gomez & Carvalho, 2000) and placed individually to hatch at 6 g l−1 artificial sea water and kept at 25°C and constant illumination. Hatchlings were isolated individually, and single-clone cultures were then established by allowing parthenogenetic proliferation. These clonal cultures were taxonomically identified as B. plicatilis using restriction fragment length polymorphism analysis (RFLP) on a fragment of the mitochondrial gene cytochrome oxidase I (COI) (Campillo et al., 2005). A total of 30–40 clones per population were established in 2017 except for CAM, which were established in 2019.
Experimental cultures
We created multiclonal laboratory cultures for each population by placing 5 females carrying asexual female eggs by clone in 2 l glass containers (30 clones × 5 = 150 individuals). These cultures were kept in the same standard conditions as the single-clone cultures but fed with microalgae at 2×105 cells ml−1. The resulting density ensures that sexual reproduction does not occur.
After 4 days the multiclonal laboratory cultures were filtered through a 30-µm Nytal mesh sieve, and 150 egg-bearing females were picked up. The collected females from two multiclonal laboratory cultures were combined in 6 l of medium and cultured under the same conditions as above but with a microalgae concentration of 1×106 cells ml−1. A total of 28 admixed experimental cultures, resulting from the combination of seven populations (Franch-Gras et al., 2017b) referred as “A” populations) with SAL and RAS (referred as “B” populations) was obtained (Fig. 1, Table 2). SAL was selected as inhabiting the most predictable environment and RAS in the intermediate level of unpredictability. Two replicates per combination were set up.
Sampling of the experimental cultures
Admixed experimental cultures were sampled on day 15, 22, and 29 (hereafter called “temporal samples”). We used a vacuum pump to extract 5 l of the water column taking care of not disturbing the bottom of the container, where most of the diapausing eggs accumulate. The remaining volume was filtered through a 30-µm Nytal mesh sieve and diapausing eggs were collected and stored in 60 g l−1 artificial sea water in the dark at 4°C in order to prevent spontaneous hatching. We then returned the 5 l of the water column to the glass container, adding 1 l of medium with microalgae at 1×106 cells ml−1. Only diapausing eggs from the temporal samples of day 15 and 29 were used for subsequent analysis. Density of diapausing eggs of day 15 and 29 was estimated in order to cover different phases of the population dynamics. This density was estimated using a particle counter (Coulter Counter, Beckman) by counting an aliquot of the sample.
DNA extraction and egg genotyping
We selected 46 healthy-looking diapausing eggs from each replicate from the temporal samples collected on days 15 and 29. DNA was extracted from each egg separately. To do this, the diapausing eggs were washed in distilled water and placed separately in 0.2 ml Eppendorf PCR tubes with 20 µl of 5 mM TE buffer and sonicated during 60 s in an Ultrasonic bath FB 15,047, Fisherbrand™ at room temperature. As the rigid chitin-like double external layer covering rotifer diapausing eggs (García-Roger et al., 2005; Denekamp et al., 2010) is sometimes resistant to breakage, the success of the sonication was inspected visually under a stereo microscope (Olympus SZX10, Japan) by searching for an open egg envelope without visible inner content of the eggs. If failed, we manually disrupted the external layer using a pipette tip and repeated the sonication. DNA was stored at − 20°C until further use.
For the identification of the diapausing eggs to inbred (AxA, BxB) or outbred (AxB) crosses we used Kompetitive allele specific PCR (KASP™, LGC Genomics, Teddington, Middlesex, UK) genotyping assays. Using available genomic information (Franch-Gras et al., 2018) for the same natural populations, we designed a set of SNPs with private or quasi-private alleles to discriminate between pairs of populations (Suppl. Table 1). DNA samples from single diapausing eggs were analysed by KASP™ genotyping assays by Biosearch Technologies (UK). We analysed 92 samples for each population combination from two temporal samples (days 15 and 29).
Due to the low amount of DNA in a single B. plicatilis egg, KASP™ genotyping often had problems in identifying the genotypes. For this reason, each sample was run twice and the following genotyping rules were adopted: (1) if both runs produced the same genotype, the resulting genotypes was assigned to the diapausing egg; (2) if only one run produced results, it was assumed as the egg genotype; and (3) in case of a discordant genotyping—either (a) different homozygotes or (b) heterozygote plus homozygote—between runs, it was assumed as an effect of allele dilution due to the low concentration of DNA and the heterozygote genotype was assigned to the egg. Notice that these rules might result in an overestimation of heterozygotes, therefore being a conservative assumption in relation to the hypothesis of reproductive isolation.
Data analysis
We calculated the percentage of all three genotypes (AxA inbred, BxB inbred, and AxB outbred) in the diapausing eggs for each temporal sample separately (14 combinations of natural populations × 2 replicates × 2 temporal samples). Fixation index (FIS) and its significance for heterozygote deficiency (uni-directional tests) was calculated using GenePop on the web (Raymond 1995, Rousset 2008). FIS ranges from − 1 (complete outbreeding) to + 1 (complete inbreeding). Before testing for heterozygote deficiency, in order to increase the power of subsequent statistical tests, we performed a chi-square test of homogeneity for the two temporal samples and the two replicates of each population combination, using the chisq.test implemented in R software (ver. 4.1.2) and pooled those samples that resulted homogeneous. Additionally, several average FIS were computed after weighting with egg abundance (see "Results"). The correlation between FIS values of the two replicates within-population combinations, as well as correlation between FIS and the different predictors (geographic, environmental and genetic pairwise distance, Table 2) was calculated using cor.test function implemented in R software (Pearson correlation, one sided). In these correlations, environmental distance was computed as the difference between the unpredictability values from each pond according to Franch-Gras et al. (2017a). Genetic distance was based on the FST value of microsatellites and φST value for the terminal repeat of the mate recognition gene mmr-b (Jezkova et al., 2022b).
Results
All 28 admixed experimental cultures produced diapausing eggs. Egg densities varied between replicates and mostly along the experimental time course. Based on these egg densities, the daily egg production rate ranged from 0.04 to 1.54 eggs ml−1 day−1 on day 15, and from 0.02 to 0.46 eggs ml−1 day−1 on day 29. In all cases, accumulated egg density on day 15 (which integrates two weeks of production) was much higher than the corresponding day 29 of sampling (one week of production) (Suppl. Table 2).
A total of 4539 SNPs were previously identified for the different populations (Franch-Gras et al., 2018), from those, only 276 SNPs had the capability to differentiate between natural populations in at least one of the 14 population combinations. We were able to design a KASP™ assay for all population combinations in our experimental design except in two cases (RASxHYB and SALxMNT), where only one and two SNPs respectively were able to differentiate between the two populations involved in the cross but gave no result in the KASP™ assays. These two population combinations were excluded from further analysis. Genotypes could be assigned to 2005 eggs (between 27 and 46 eggs per replicate and population combination). Frequencies of genotypes are shown in Fig. 2. Genotyping rules (see "Material and Methods") had to be applied to 50.4% of the genotypes as they presented inconsistencies among KASP™ runs. From those, 72.0% were classified as heterozygotes. The percentage of manually assigned heterozygotes varied among population combinations (interquartile range: 49.9 and 90.7%). Deficiency of heterozygotes was observed in 26 out of 48 samples. Temporal samples and replicates were pooled in 7 combinations (RASxAYA, RASxHYC, RASxMNT, RASxPET and SALxHYB, SALxPET, SALxTUR) after testing for homogeneity of the genotypic frequencies. After pooling homogeneous samples, significant deficiency of heterozygosity was found in four population combinations (RASxMNT, RASxPET, SALxHYB, SALxTUR; pooled replicate and dates), and in the 15-day temporal samples of both replicates of RASxTUR. That is, we found significant heterozygote deficiency in 6 out of 27 tests, involving 5 out of 12 population combinations.
Percentage of diapausing eggs genotypes produced in each population combination for two temporal samples (day 15 and day 29) and two replicates (a and b). Those population combinations with homogeneous samples were pooled for analysis and highlighted in black. Stars indicate significant deficit of heterozygotes, black horizontal lines accounts for significant deficit of heterozygotes in pooled replicates and samples
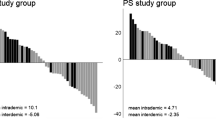
The FIS values of the 48 samples ranged from − 0.84 to + 0.54. Correlation of FIS values between the two replicates were highly significant (P < 0.001 for both days 15 and 29). Six out of 12 population combinations showed positive average FIS values (Fig. 3, average over population combinations: 0.062). A trend of decreasing FIS was observed between day 15 and day 29 in 10 out of 12 population combinations. When comparing the FIS separately for each temporal sample (day 15 vs. day 29), average FIS on day 15 was positive (0.112), while on day 29 was negative (− 0.141). FIS (either per population combination or for each temporal sample) did not show significant correlation with any of the predictors (environmental, geographic or genetic distance) (Suppl. Figure 1).
FIS values for each population combination and replicate for the two temporal samples (day 15 and day 29). Black dashed line is averaged over replicates. The number within each panel is FIS averaged over temporal samples and replicates. The area above the line corresponds to within-population mating preferences and below the line to among population mating preferences
Discussion
In this study we have investigated the evolution of an incipient reproductive isolation among populations of B. plicatilis from geographically close ponds that differ in their environmental conditions. We selected nine populations locally adapted to water regime unpredictability (Franch-Gras et al., 2017b). We have produced diapausing eggs from 12 population combinations and found signs of assortative mating (deficit of heterozygotes) in five of them. The emergence of reproductive isolation in the presence of gene flow is not easy to evolve as gene flow tends to blur divergence and because mating recognition systems are under stabilising selection (Lambert et al., 1982; Butlin et al., 1985). Therefore, our observation of deficit of heterozygotes in more than one third of our experimental cultures seems to us biologically significant.
The index of inbreeding (weighted FIS) was positive in eight out of 12 population combinations, providing signatures for outcross avoidance. The overall FIS value was very varying (FIS average 0.062; range − 0.84 to 0.54). The most inbred population combination in this study was SALxHYB, with an average FIS of 0.25. Interestingly, in an independent previous study on mating behaviour, these populations also showed a significant preference for within-population mating (Jezkova et al., 2022a), largest differences in environmental predictability (Franch-Gras et al., 2017a) and each population belongs to a different mmr-b haplotype group (Jezkova et al., 2022b).
Regarding those populations that did not show a deficit of heterozygotes (4 out of 12), different explanations could be invoked. Our results could be due to an actual lack of reproductive barrier between these populations. On the other hand, the experimental setup could have affected the outcome increasing the number of heterozygotes found in several ways. First, the amount of DNA obtained from a single egg is small, and this can affect the genotyping by KASP™ analyses resulting in random loss of one allele. KASP™ was run twice for each egg and every inconsistency (one different homozygote each time) was assigned as heterozygote. By doing this, we may have biased our data against our hypothesis of heterozygote deficit. Second, by admixing populations at high concentrations (150 + 150 females in 6 l of media) we may have increased the probability of encounters and forced random mating, overcoming the effect of the pre-reproductive barrier found in Jezkova et al. (2022a). In the wild, this equifrequent assemblage would not occur. Third, as explained below, asynchronous timing of sexual reproduction may explain events of heterozygote excess.
Whereas average FIS, either globally or in eight out of 12 population combinations, was positive indicating a tendency towards within-population crossing, this index decreased between day 15 (global FIS 0.112) and day 29 (global FIS − 0.141, with low effect due to de decreased egg production). Two different biological factors could contribute to this pattern. One factor may be the environment deterioration between day 15 and day 29 (e.g. accumulation of metabolic compounds or debris). This deterioration might be appreciated by rotifers as the end of the growing season and lead to lower partner discrimination at later stages (better a bad mate than no mate). This would imply FIS to tend to zero. The other putative factor is related to different timing of induction of sexual reproduction. Asynchrony of reproductive cycles has been reported as one of the important components of reproductive isolation in many species (Wu et al., 2021). In our study system, natural populations are locally adapted to environmental unpredictability by adjusting their density threshold for sex induction, that is, some populations induce sex earlier than others (Franch-Gras et al., 2017b). Accordingly, when the later-inducer population starts producing sexual females, males from the earlier-inducer population are already copulating with them, increasing the number of outbred crosses. Preliminary results from a model simulation (Serra, unpublished) suggest that this coupling might explain a period of heterozygote deficiency followed by a period of heterozygote excess.
Previous studies suggest a tendency towards pre-reproductive isolation related to environmental distance (Jezkova et al., 2022a). In this study, however, despite signatures for within-population crossing, our hypothesis of a stronger deficit of heterozygotes in those populations that are more ecologically distant was not observed. The tendency of populations to avoid interbreeding was not related to environmental distance, nor to geographic or genetic distance. Lack of correlation with genetic distance occurred for neutral markers (microsatellites) and mmr-b gene, which is involved in rotifer mate recognition (see Jezkova et al., 2022b). We do not consider this lack of evidence definitive. Firstly, our study was limited to 14 population combinations sampled at a regional scale (240 km2), so lack of correlation could be due to low statistical power in order to detect a small effect. Secondly, as previously pointed, experimental factors and inherent biological characteristic of our populations may have altered the frequency of heterozygotes in our experiments, and thus, altering the relationship among genetic and environmental distance. The natural populations studied here belong to two aminoacidic haplotype groups of the mmr-b gene (Jezkova et al., 2022b). SAL and TUR are in one group and the rest in the other. Correlations of FIS with φST of mmr-b in population combinations involving SAL, but excluding TUR with which there is no genetic distance, resulted in a positive correlation (P = 0.05, Suppl. Fig. 1) on the edge of the statistical significance. This suggests a role of the mating recognition gene in population divergence but also the high complexity of mating discrimination and behavioural isolation, where a panoply of biotic and abiotic factors might be at work. Indeed, our experimental setup was based on oversimplified populations formed by an equal admixture of differentially adapted individuals in a homogeneous environment. Moreover, the involvement of additional genes controlling successful reproduction (Snell, 2011; Hanson et al., 2013) cannot be discarded.
Zooplankton populations in nature are genetically structured despite their high dispersal capacity (De Meester, 1996; De Meester et al., 2002). This paradox has been explained as the result of persistent founder effects and local adaptation (De Meester et al., 2002; Montero-Pau et al., 2018). There is evidence for local adaptation in cladocerans, rotifers and other zooplanktonic groups (Fernández et al., 2020; Tangwancharoen et al., 2020; White et al., 2022). In this scenario, the evolution of a reproductive barrier among locally adapted populations is advantageous. Our study points in this direction. However, a trade off may exist between preserving favourable gene combinations that are advantageous in certain habitats and undesirable negative consequences of inbreeding (Tortajada et al., 2009; Cannon, 2021). This trade off might weaken the reproductive barrier, resulting in a gradient of reproductive isolation. In this study we have found signatures of incipient reproductive isolation in some populations of B. plicatilis from geographical proximity with putative migration among them. The fact that we worked at an intraspecific level and controlling only a limited set of factors is consistent with not finding clear cuts for isolation. Given that the evolution of reproductive isolation and speciation are rare historical events in nature, finding partial isolation within some of the populations studies seems to us biologically significant. By finding partial within-species reproductive isolation between populations, our results make worthy further investigation on the role of local adaptation in shaping the diversification of small zooplankton populations.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Butlin, R., 1987. Speciation by reinforcement. Trends in Ecology and Evolution 2: 8–13.
Butlin, R. K., G. M. Hewitt & S. F. Webb, 1985. Sexual selection for intermediate optimum in Chorthippus brunneus (Orthoptera: Acrididae). Animal Behaviour 33: 1281–1292.
Campillo, S., E. M. García-Roger, D. Martínez-Torres & M. Serra, 2005. Morphological stasis of two species belonging to the L-morphotype in the Brachionus plicatilis species complex. Hydrobiologia 546: 181–187.
Cannon, C. H., 2021. Is speciation an unrelenting march to reproductive isolation? Molecular Ecology 30: 4349–4352.
Carmona, M. J., M. Serra & M. R. Miracle, 1993. Relationships between mixis in Brachionus plicatilis and preconditioning of culture medium by crowding. Hydrobiologia 255: 145–152.
Chin, T. A., C. E. Cáceres & M. E. Cristescu, 2019. The evolution of reproductive isolation in Daphnia. BMC Evolutionary Biology 19: 1–15.
Coyne, J. & H. A. Orr, 1989. Patterns of speciation in drosophila. Evolution 43: 362–381.
Coyne, J. A. & H. A. Orr, 2004. Speciation, Sinauer Associates, Sunderland
De Meester, L., 1996. Local genetic differentiation and adaptation in freshwater zooplankton populations: patterns and processes. Écoscience 3: 385–399.
De Meester, L., A. Gomez, B. Okamura & K. Schwenk, 2002. The monopolization hypothesis and the dispersal-gene flow paradox in aquatic organisms. Acta Oecologica 23: 121–135.
Declerck, S. A. J. & S. Papakostas, 2016. Monogonont rotifers as model systems for the study of micro-evolutionary adaptation and its eco-evolutionary implications. Hydrobiologia 796: 131–144.
Denekamp, N. Y., K. Suga, A. Hagiwara, R. Reinhardt & E. Lubzens, 2010. A role for molecular studies in unveiling the pathways for formation of rotifer resting eggs and their survival during dormancy. Topics in Current Genetics 21: 109–132.
Feder, J. L., S. M. Flaxman, S. P. Egan, A. A. Comeault & P. Nosil, 2013. Geographic mode of speciation and genomic divergence. Annual Review of Ecology, Evolution, and Systematics 44: 73–97.
Fernández, C. E., M. Campero, G. Bianco, M. T. Ekvall, D. Rejas, C. B. Uvo & L. A. Hansson, 2020. Local adaptation to UV radiation in zooplankton: a behavioral and physiological approach. Ecosphere 11: e03081.
Franch-Gras, L., E. M. García-Roger, B. Franch, M. J. Carmona & M. Serra, 2017a. Quantifying unpredictability: a multiple-model approach based on satellite imagery data from Mediterranean ponds. PLOS ONE 12: e0187958.
Franch-Gras, L., E. M. García-Roger, M. Serra & M. José Carmona, 2017b. Adaptation in response to environmental unpredictability. Proceedings of the Royal Society B: Biological Sciences 284: 20170427.
Franch-Gras, L., C. Hahn, E. M. García-Roger, M. J. Carmona, M. Serra & A. Gomez, 2018. Genomic signatures of local adaptation to the degree of environmental predictability in rotifers. Scientific Reports 8: 1–14.
García-Roger, E. M., M. J. Carmona & M. Serra, 2005. Deterioration patterns in diapausing egg banks of Brachionus (Müller, 1786) rotifer species. Journal of Experimental Marine Biology and Ecology 314: 149–161.
Gilbert, J. J., 2007. Timing of Diapause in Monogonont Rotifers: Mechanisms and Strategies. In Alekseev, V. R., B. T. de Stasio & J. J. Gilbert (eds), Diapause in Aquatic Invertebrates Theory and Human Use. Monographiae Biologicae, Springer, Dordrecht.
Gomez, A. & G. R. Carvalho, 2000. Sex, parthenogenesis and genetic structure of rotifers: microsatellite analysis of contemporary and resting egg bank populations. Molecular Ecology 9: 203–214.
Gomez, A. & M. Serra, 1995. Behavioral reproductive isolation among sympatric strains of Brachionus plicatilis Müller 1786: insights into the status of this taxonomic species. Hydrobiologia 313–314: 111–119.
Hanson, S. J., C. P. Stelzer, D. B. M. Welch & J. M. Logsdon, 2013. Comparative transcriptome analysis of obligately asexual and cyclically sexual rotifers reveals genes with putative functions in sexual reproduction, dormancy, and asexual egg production. BMC Genomics 14: 1–17.
Jezkova, I., R. Ortells, J. Montero-Pau & M. Serra, 2022a. Insight into incipient reproductive isolation in diverging populations of a rotifer Brachionus plicatilis. Hydrobiologia 849: 3299–3311.
Jezkova, I., M. Serra, R. Ortells & J. Montero, 2022b. Genetic variability of the mating recognition gene in populations of Brachionus plicatilis. Diversity 14: 155.
Kordbacheh, A., A. N. Shapiro & E. J. Walsh, 2019. Reproductive isolation, morphological and ecological differentiation among cryptic species of Euchlanis dilatata, with the description of four new species. Hydrobiologia 844: 221–242.
Kordbacheh, A., H. Rahimian & D. Fontaneto, 2023. Mechanisms of reproductive isolation among cryptic species in monogonont rotifers. Hydrobiologia. https://doi.org/10.1007/s10750-022-05131-0.
Lambert, D. M., P. D. Kingett & E. Slooten, 1982. Intersexual selection: the problem and a discussion of the evidence. Evolutionary Theory 6: 67–78.
Mayr, E., 1963. Animal Species and Evolution, Belknap Press of Harvard University, Cambridge
Montero-Pau, J., A. Gomez & M. Serra, 2018. Founder effects drive the genetic structure of passively dispersed aquatic invertebrates. PeerJ 6: e6094.
Nosil, P., 2013. Degree of sympatry affects reinforcement in Drosophila. Evolution 67: 868–872.
Raymond M. & Rousset F, 1995. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. Journal of Heredity 86: 248–249.
Rousset, F., 2008. Genepop’007: A complete reimplementation of the Genepop software for Windows and Linux. Molecular Ecology Resources 8: 103–106.
Schröder, T. & E. J. Walsh, 2007. Cryptic speciation in the cosmopolitan Epiphanes senta complex (Monogononta, Rotifera) with the description of new species. Hydrobiologia 593: 129–140.
Snell, T. W., 2011. A review of the molecular mechanisms of monogonont rotifer reproduction. Hydrobiologia 662: 89–97.
Stelzer, C. P. & T. W. Snell, 2003. Induction of sexual reproduction in Brachionus plicatilis (Monogononta, Rotifera) by a density-dependent chemical cue. Limnology and Oceanography 48: 939–943.
Suatoni, E., S. Vicario, S. Rice, T. Snell & A. Caccone, 2006. An analysis of species boundaries and biogeographic patterns in a cryptic species complex: the rotifer-Brachionus plicatilis. Molecular Phylogenetics and Evolution 41: 86–98.
Tangwancharoen, S., B. X. Semmens & R. S. Burton, 2020. Allele-specific expression and evolution of gene regulation underlying acute heat stress response and local adaptation in the copepod Tigriopus californicus. Journal of Heredity 111: 539–547.
Tortajada, A. M., M. J. Carmona & M. Serra, 2009. Does haplodiploidy purge inbreeding depression in rotifer populations? PLoS ONE 4: e8195.
Tregenza, T., 2002. Divergence and reproductive isolation in the early stages of speciation. Genetica 116: 291–300.
Westram, A. M., S. Stankowski, P. Surendranadh & N. Barton, 2022. What is reproductive isolation? Journal of Evolutionary Biology 35: 1143–1164.
White, N. J., A. P. Beckerman, R. R. Snook, M. A. Brockhurst, R. K. Butlin & I. Eyres, 2022. Experimental evolution of local adaptation under unidimensional and multidimensional selection. Current Biology 32: 1310-1318.e4.
Wu, H., H. Wang & S. Ding, 2021. Reproductive biology and annual reproductive cycles of two sympatric lineages of Bostrychus sinensis with a natural habitat on southeastern coast of China. Animal Reproduction Science 232: 106821.
Yukilevich, R., 2012. Asymmetrical patterns of speciation uniquely support reinforcement in Drosophila. Evolution 66: 1430–1446.
Zhang, W. & S. A. J. Declerck, 2022a. Reduced fertilization constitutes an important prezygotic reproductive barrier between two sibling species of the hybridizing Brachionus calyciflorus species complex. Hydrobiologia 849: 1701–1711.
Zhang, W. & S. A. J. Declerck, 2022b. Intrinsic postzygotic barriers constrain cross-fertilisation between two hybridising sibling rotifer species of the Brachionus calyciflorus species complex. Freshwater Biology 67: 240–249.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. The research was supported by a grant from the Valencian Conselleria de Educación, Investigación, Cultura y Deporte (Grant No. AICO/2020/013), a Grant from the Spanish Ministerio de Ciencia e Innovación (Grant No. CIN CGL2015-65422-P) and the European Grant “ERDF A way of making Europe”. IJ was supported by a grant from the Valencian Consellería de Educación, Investigación, Cultura y Deporte (Grant No. GRISOLIAP/2017/096).
Author information
Authors and Affiliations
Contributions
IJ, RO and MS contributed to the study conception and design. Material preparation and data collection were performed by IJ and JM-P. Data analysis were carried by IJ, JM-P, RO and MS. The first draft of the manuscript was written by IJ and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Guest editors: Maria Špoljar, Diego Fontaneto, Elizabeth J. Walsh & Natalia Kuczyńska-Kippen / Diverse Rotifers in Diverse Ecosystems
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jezkova, I., Montero-Pau, J., Ortells, R. et al. Development of reproductive barriers in sympatry. Hydrobiologia 851, 2927–2936 (2024). https://doi.org/10.1007/s10750-023-05233-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-023-05233-3