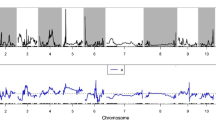
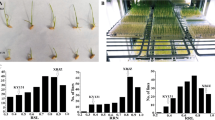
Abstract
Main Conclusion: The major QTL qATS1 for rice seedling alkali tolerance was identified by QTL-seq and QTL mapping, and further confirmed that OsMYB305 regulates plant alkali tolerance by affecting the transport of Na+ and K+ in roots and seedlings through qRT-PCR and functional characterization. Rice is a crop sensitive to saline and alkaline stress. However, the scarcity of alkali tolerance genes in rice has resulted in a large number of cultivated rice or germplasm resources that are difficult to cultivate in saline fields. To clone new alkali tolerance genes, we systematically characterized the number of days of seedling survival and sodium and potassium ion concentrations under alkali stress using 840 F2:3 individuals generated from a cross between Hajingdao8 and Tengxi144. Using QTL-seq technology and QTL mapping based on KASP markers, qATS1 was localized to the 77.15 Kb interval on chromosome 1. A MYB gene, OsMYB305, was identified to strongly respond to alkali stress by gene function annotation, mutation detection, and qRT-PCR analysis of this interval. Knockdown of OsMYB305 in HJD8 resulted in a massive transfer of Na+ and K+ from the mutant roots to the leaves, leading to the death of osmyb305 leaves after 18 days of alkali stress. In conclusion, this study identified OsMYB305 as an alkali tolerance gene, and the variation of 19 SNPs in the promoter region of OsMYB305 and 4 SNPs on exons can provide a theoretical basis for future search of molecular mechanisms upstream and downstream of OsMYB305 to regulate alkali tolerance and molecular design breeding.





Similar content being viewed by others
Data availability
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession numbers can be found below: NCBI SRA: SRR27393491, SRR27393492, SRR27393493, and SRR27393494.
References
Asano T, Hayashi N, Kobayashi M, Aoki N, Miyao A, Mitsuhara I, Ichikawa H, Komatsu S, Hirochika H, Kikuchi S, Ohsugi R (2012) A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J 69(1):26–36. https://doi.org/10.1111/j.1365-313X.2011.04766.x
Chakraborty K, Chattaopadhyay K, Nayak L, Ray S, Yeasmin L, Jena P, Gupta S, Mohanty SK, Swain P, Sarkar RK (2019) Ionic selectivity and coordinated transport of na + and K + in flag leaves render differential salt tolerance in rice at the reproductive stage. Planta 250(5):1637–1653. https://doi.org/10.1007/s00425-019-03253-9
Chen H, Zhang Q, Cai H, Xu F (2017) Ethylene mediates Alkaline-Induced Rice Growth Inhibition by negatively regulating plasma membrane H(+)-ATPase activity in roots. Front Plant Sci 8:1839. https://doi.org/10.3389/fpls.2017.01839
Chuamnakthong S, Nampei M, Ueda A (2019) Characterization of na(+) exclusion mechanism in rice under saline-alkaline stress conditions. Plant Sci 287:110171. https://doi.org/10.1016/j.plantsci.2019.110171
De Leon TB, Linscombe S, Subudhi PK (2016) Molecular Dissection of Seedling Salinity Tolerance in Rice (Oryza sativa L.) Using a High-Density GBS-Based SNP Linkage Map. Rice (New York, NY) 9 (1):52. https://doi.org/10.1186/s12284-016-0125-2
Guo M, Wang R, Wang J, Hua K, Wang Y, Liu X, Yao S (2014) ALT1, a Snf2 family chromatin remodeling ATPase, negatively regulates alkaline tolerance through enhanced defense against oxidative stress in rice. PLoS ONE 9(12):e112515. https://doi.org/10.1371/journal.pone.0112515
Hill JT, Demarest BL, Bisgrove BW, Gorsi B, Su Y-C, Yost HJ (2013) MMAPPR: Mutation Mapping Analysis Pipeline for pooled RNA-seq. Genome Res 23(4):687–697. https://doi.org/10.1101/gr.146936.112
Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23(15):1805–1817. https://doi.org/10.1101/gad.1812409
Kumar S, Agarwal S, Ranvijay (2019) Fast and memory efficient approach for mapping NGS reads to a reference genome. J Bioinf Comput Biol 17(2). https://doi.org/10.1142/s0219720019500082
Lei L, Cao L, Ding G, Zhou J, Luo Y, Bai L, Xia T, Chen L, Wang J, Liu K, Lei Q, Xie T, Yang G, Wang X, Sun S, Lai Y (2023) OsBBX11 on qSTS4 links to salt tolerance at the seeding stage in Oryza sativa L. Ssp. Japonica Front Plant Sci 8:14:1139961. https://doi.org/10.3389/fpls.2023.1139961
Li Q, Yang A, Zhang WH (2016) Efficient acquisition of iron confers greater tolerance to saline-alkaline stress in rice (Oryza sativa L). J Exp Bot 67(22):6431–6444. https://doi.org/10.1093/jxb/erw407
Li N, Sun J, Wang J, Liu H, Zheng H, Yang L, Liang Y, Li X, Zou D (2017) QTL analysis for alkaline tolerance of rice and verification of a major QTL. Plant Breeding 136(6):881–891. https://doi.org/10.1111/pbr.12539
Li N, Zheng H, Cui J, Wang J, Liu H, Sun J, Liu T, Zhao H, Lai Y, Zou D (2019) Genome-wide association study and candidate gene analysis of alkalinity tolerance in japonica rice germplasm at the seedling stage. Rice 12(1):24. https://doi.org/10.1186/s12284-019-0285-y
Li X, Zheng H, Wu W, Liu H, Wang J, Jia Y, Li J, Yang L, Lei L, Zou D, Zhao H (2020) QTL mapping and Candidate Gene Analysis for Alkali Tolerance in Japonica Rice at the bud stage based on linkage mapping and genome-wide Association study. Rice (New York NY) 13(1):48. https://doi.org/10.1186/s12284-020-00412-5
Liang JL, Qu YP, Yang CG, Ma XD, Cao GL, Zhao ZW, Zhang SY, Zhang T, Han LZ (2015) Identification of QTLs associated with salt or alkaline tolerance at the seedling stage in rice under salt or alkaline stress. Euphytica 201(3):441–452. https://doi.org/10.3864/j.issn.0578-1752.2017.10.001
Liu L, Wang B (2021) Protection of Halophytes and their uses for cultivation of saline-alkali soil in China. Biology 10(5). https://doi.org/10.3390/biology10050353
Liu K, Wang J, Gao S, Liu Q, Shan L, Guo J, Lei L, Bian J (2023) Combined QTL-seq and QTL mapping strategies identify genes related to alkalinity tolerance in rice seedlings. Plant Growth Regul 101(3):781–792. https://doi.org/10.1007/s10725-023-01056-3
Lu X, Min W, Shi Y, Tian L, Li P, Ma T, Zhang Y, Luo C (2022) Exogenous melatonin alleviates alkaline stress by removing reactive oxygen species and promoting antioxidant defence in Rice Seedlings. Front Plant Sci 13:849553. https://doi.org/10.3389/fpls.2022.849553
Luo X, Deng H, Wang P, Zhang X, Li C, Li C, Tan J, Wu G, Wang Y, Cheng Q (2020) Genetic analysis of germinating ability under alkaline and neutral salt stress by a high-density bin genetic map in rice. Euphytica 216(7):1–12. https://doi.org/10.1007/s10681-020-02623-9
Lv BS, Ma HY, Li XW, Wei LX, Liang ZW (2015) Proline Accumulation is not correlated with saline-alkaline stress tolerance in Rice Seedlings. Agron J 107(1):51
Magwene PM, Willis JH, Kelly JK (2011) The statistics of bulk segregant analysis using next generation sequencing. PLoS Comput Biol 7(11):e1002255. https://doi.org/10.1371/journal.pcbi.1002255
Mansfeld BN, Grumet R (2018) QTLseqr: an R Package for Bulk Segregant Analysis with Next-Generation sequencing. Plant Genome 11(2). https://doi.org/10.3835/plantgenome2018.01.0006
Mittal D, Chakrabarti S, Sarkar A, Singh A, Grover A (2009) Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol Biochem 47(9):785–795. https://doi.org/10.1016/j.plaphy.2009.05.003
Moradi F, Ismail AM (2007) Responses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Ann Bot 99(6):1161–1173. https://doi.org/10.1093/aob/mcm052
Ontoy JC, Shrestha B, Karki HS, Barphagha I, Angira B, Famoso A, Ham JH (2023) Genetic characterization of the partial Disease Resistance of Rice to bacterial panicle blight and sheath blight by combined QTL linkage and QTL-seq analyses. Plants (Basel) 12(3):559. https://doi.org/10.3390/plants12030559
Prakash NR, Lokeshkumar BM, Rathor S, Warraich AS, Yadav S, Vinaykumar NM, Dushynthkumar BM, Krishnamurthy SL, Sharma PC (2022) Meta-analysis and validation of genomic loci governing seedling and reproductive stage salinity tolerance in rice. Physiol Plant 174(1):e13629. https://doi.org/10.1111/ppl.13629
Qi D, Guo G, Lee MC, Zhang J, Cao G, Zhang S, Suh SC, Zhou Q, Han L (2008) Identification of quantitative trait loci for the dead leaf rate and the seedling dead rate under alkaline stress in rice. J Genet Genomics = Yi Chuan Xue bao 35(5):299–305. https://doi.org/10.1016/s1673-8527(08)60043-0
Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, Blée E, Mongrand S, Domergue F, Roby D (2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20(3):752–767. https://doi.org/10.1105/tpc.107.054858
Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37(10):1141–1146. https://doi.org/10.1038/ng1643
Sandhu N, Singh J, Singh G, Sethi M, Singh MP, Pruthi G, Raigar OP, Kaur R, Kaur R, Sarao PS, Lore JS, Singh UM, Dixit S, Sagare DB, Singh SP, Satturu V, Singh VK, Kumar A (2022) Development and validation of a novel core set of KASP markers for the traits improving grain yield and adaptability of rice under direct-seeded cultivation conditions. Genomics 114(2):110269. https://doi.org/10.1016/j.ygeno.2022.110269
Singh K, Foley RC, Oñate-Sánchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5(5):430–436. https://doi.org/10.1016/s1369-5266(02)00289-3
Singh L, Coronejo S, Pruthi R, Chapagain S, Bhattarai U, Subudhi PK (2022a) Genetic dissection of alkalinity tolerance at the Seedling Stage in Rice (Oryza sativa) using a high-resolution linkage map. Plants (Basel Switzerland) 11(23). https://doi.org/10.3390/plants11233347
Singh L, Coronejo S, Pruthi R, Chapagain S, Subudhi PK (2022b) Integration of QTL mapping and whole genome sequencing identifies candidate genes for alkalinity tolerance in Rice (Oryza sativa). Int J Mol Sci 23(19). https://doi.org/10.3390/ijms231911791
Smolen GA, Pawlowski L, Wilensky SE, Bender J (2002) Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis. Genetics 161(3):1235–1246. https://doi.org/10.1093/genetics/161.3.1235
Sugimoto K, Takeda S, Hirochika H (2000) MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell 12(12):2511–2528. https://doi.org/10.1105/tpc.12.12.2511
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74(1):174–183. https://doi.org/10.1111/tpj.12105
Takehisa H, Shimodate T, Fukuta Y, Ueda T, Yano M, Yamaya T, Kameya T, Sato T (2004) Identification of quantitative trait loci for plant growth of rice in paddy field flooded with salt water. Field Crops Res 89(1):85–95. https://doi.org/10.1016/j.fcr.2004.01.026
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16):e164. https://doi.org/10.1093/nar/gkq603
Wang H, Lin X, Cao S, Wu Z (2015a) Alkali tolerance in rice (Oryza sativa L.): growth, photosynthesis, nitrogen metabolism, and ion homeostasis. Photosynthetica 53(1):55–65
Wang R, Jing W, Xiao L, Jin Y, Shen L, Zhang W (2015b) The Rice High-Affinity Potassium Transporter1;1 is involved in Salt Tolerance and regulated by an MYB-Type transcription factor. Plant Physiol 168(3):1076–1090. https://doi.org/10.1104/pp.15.00298
Wang J, Nan N, Li N, Liu Y, Wang TJ, Hwang I, Liu B, Xu ZY (2020) A DNA methylation Reader-Chaperone Regulator-transcription factor Complex activates OsHKT1;5 expression during salinity stress. Plant Cell 32(11):3535–3558. https://doi.org/10.1105/tpc.20.00301
Wei LX, Lv BS, Wang MM, Ma HY, Yang HY, Liu XL, Jiang CJ, Liang ZW (2015) Priming effect of abscisic acid on alkaline stress tolerance in rice (Oryza sativa L.) seedlings. Plant Physiol Biochem 90:50–57. https://doi.org/10.1016/j.plaphy.2015.03.002
Xiong H, Li J, Liu P, Duan J, Zhao Y, Guo X, Li Y, Zhang H, Ali J, Li Z (2014) Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 9(3):e92913. https://doi.org/10.1371/journal.pone.0092913
Yang A, Dai X, Zhang WH (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 63(7):2541–2556. https://doi.org/10.1093/jxb/err431
Yeo AR, Yeo ME, Flowers SA, Flowers TJ (1990) Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theor Appl Genet 79(3):377–384. https://doi.org/10.1007/BF01186082
Yokotani N, Ichikawa T, Kondou Y, Iwabuchi M, Matsui M, Hirochika H, Oda K (2013) Role of the rice transcription factor JAmyb in abiotic stress response. J Plant Res 126(1):131–139. https://doi.org/10.1007/s10265-012-0501-y
Zhang S, Haider I, Kohlen W, Jiang L, Bouwmeester H, Meijer AH, Schluepmann H, Liu CM, Ouwerkerk PB (2012) Function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol Biol 80(6):571–585. https://doi.org/10.1007/s11103-012-9967-1
Zhang H, Liu XL, Zhang RX, Yuan HY, Wang MM, Yang HY, Ma HY, Liu D, Jiang CJ, Liang ZW (2017) Root damage under Alkaline stress is Associated with reactive oxygen species Accumulation in Rice (Oryza sativa L). Front Plant Sci 8:1580. https://doi.org/10.3389/fpls.2017.01580
Zhang C, Zhang J, Liu H, Qu X, Wang J, He Q, Zou J, Yang K, Le J (2022) Transcriptomic analysis reveals the role of FOUR LIPS in response to salt stress in rice. Plant Mol Biol 110(1–2):37–52. https://doi.org/10.1007/s11103-022-01282-9
Zhao M, Wang Y, He N, Pang X, Wang L, Ma Z, Tang Z, Gao H, Zhang L, Fu L, Wang C, Liu J, Zheng W (2022) QTL detection for Rice Grain length and fine mapping of a novel locus qGL6.1. 15(1):60 Rice (New York, NY). https://doi.org/10.1186/s12284-022-00606-z
Zhu F, Wang K, Li D, Liu Z, Li M, Wang Z, Li X, Lan X, Guan Q (2022) OsSAP6 positively regulates Soda saline-alkaline stress tolerance in Rice. Rice (New York NY) 15(1):69. https://doi.org/10.1186/s12284-022-00616-x
Acknowledgements
The authors would like to thank all the reviewers who participated in the review and the Editor for their linguistic assistance during the preparation of this manuscript.
Funding
This research was funded by the Harbin Science and Technology Tackling Program, Heilongjiang Province, China ( GJ2021TZ002007 ).
Author information
Authors and Affiliations
Contributions
Conceptualization, QT Y.; methodology, CX L and BC C.; software and data curation, FS B; writing—original draft preparation, CX L; writing—review and editing, CX L and QT Y. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Institutional review board statement
Not applicable.
Informed consent statement
Not applicable.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, C., Chen, B., Bu, F. et al. OsMYB305 on qATS1 positively regulates alkalinity tolerance at the seedling stage in japonica rice. Plant Growth Regul (2024). https://doi.org/10.1007/s10725-024-01144-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10725-024-01144-y