Abstract
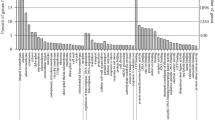
Underutilised legume Psophocarpus tetragonolobus (L.) DC. accumulates various degrees of proanthocyanidin (PA) or condensed tannin (CT) on its seed coat. Screening for PA content among various lines of P. tetragonolobus through the vanillin-HCl assay identified two contrasting lines of P. tetragonolobus (HPW and LPW). HPW contained 59.23 mg/g of total PA, with catechin and epigallocatechin gallate monomer being found among the monomers identified in HPLC; the LPW line contained 8.68 mg/g of PA in its leaves. Comparative miRNA profiling of the leaf tissues of contrasting PA lines of P. tetragonolobus revealed a total of 139 mature miRNAs along with the identification of isoforms and novel miRNAs. Differentially expressed known miRNAs, e.g., miR156, miR396, miR4414b, miR4416c, miR894, miR2111, and miR5139, were validated through qRT-PCR. Target prediction of the identified miRNAs show possible involvement of certain miRNAs like miR396, miR4414b and miR862 in PA biosynthesis in P. tetragonolobus. Hence this study can provide a basis for understanding the role of miRNAs in regulation of PA biosynthesis.







Similar content being viewed by others
Data Availability
The generated data used in this study has been submitted in the public domain database of NCBI (SRA Accession No: SRR15115657 to SRR15115660) https://www.ncbi.nlm.nih.gov. The data will be made available publicly after publication of the manuscript.
References
Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR (2003) The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J 35:624–636
Alptekin B, Akpinar BA, Budak H (2016) A comprehensive prescription for plant miRNA identification. Front Plant Sci 7:2058
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Barbehenn RV, Peter Constabel C (2011) Tannins in plant-herbivore interactions. Phytochemistry 72:1551–1565
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102:11928–11933
Beecher GR (2004) Proanthocyanidins: biological activities associated with human health. Pharm Biol 42:2–20
Bhat KV, Mondal TK, Gaikwad AB, Kole PR, Chandel G, Mohapatra T (2020) Genome-wide identification of drought-responsive miRNAs in grass pea (Lathyrus sativus L.). Plant Gene 21:100210
Brillouet JM, Romieu C, Schoefs B, Solymosi K, Cheynier V, Fulcrand H, Verdeil JL, Conejero G (2013) The tannosome is an organelle forming condensed tannins in the chlorophyllous organs of tracheophyta. Ann Botany 112:1003–1014
Chaudhary V, Jangra S, Yadav NR (2022) In silico identification of miRNAs and their targets in Cluster Bean for their role in development and physiological responses. Front Genet 13:930113
Chen J, Zheng Y, Qin L, Wang Y, Chen L, He Y, Fei Z, Lu G (2016) Identification of miRNAs and their targets through high-throughput sequencing and degradome analysis in male and female Asparagus officinalis. BMC Plant Biol 16:1–19
Chen W, Xiong Y, Xu L, Zhang Q, Luo Z (2017) An integrated analysis based on transcriptome and proteome reveals deastringency-related genes in CPCNA persimmon. Sci Rep 7:44671
Dai X, Zhuang Z, Zhao PX (2018) psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res 46:W49–W54
Dai Z, Tan J, Zhou C, Yang X, Yang F, Zhang S, Sun S, Miao X, Shi Z (2019) The OsmiR396–Os GRF 8–OsF3H-flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa). Plant Biotechnol J 17:1657–1669
DeBoer K, Melser S, Sperschneider J, Kamphuis LG, Garg G, Gao LL, Frick K, Singh KB (2019) Identification and profiling of narrow-leafed lupin (Lupinus angustifolius) microRNAs during seed development. BMC Genom 20:135
Ding T, Tomes S, Gleave AP, Zhang H, Dare AP, Plunkett B, Espley RV, Luo Z, Zhang R, Allan AC et al (2022) microRNA172 targets APETALA2 to regulate flavonoid biosynthesis in apple (Malus domestica). Hortic Res. https://doi.org/10.1093/hr/uhab007
Duodu KG, Dowell FE (2019) Sorghum and millets: quality management systems. Sorghum and millets. Elsevier, Amsterdam, pp 421–442
Fraga-Corral M, Garcia-Oliveira P, Pereira AG, Lourenco-Lopes C, Jimenez-Lopez C, Prieto MA, Simal-Gandara J (2020) Technological application of tannin-based extracts. Molecules 25:614
Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N (2012) miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40:37–52
Gao Z, Nie J, Wang H (2021) MicroRNA biogenesis in plant. Plant Growth Regul 93:1–12
Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23:1512–1522
Gul Z, Barozai MYK, Din M (2017) In-silico based identification and functional analyses of miRNAs and their targets in Cowpea (Vigna unguiculata L.). Aims Genet 4:138–165
Gupta OP, Karkute SG, Banerjee S, Meena NL, Dahuja A (2017) Contemporary understanding of miRNA-based regulation of secondary metabolites biosynthesis in plants. Front Plant Sci 8:374
He F, Pan QH, Shi Y, Duan CQ (2008) Biosynthesis and genetic regulation of proanthocyanidins in plants. Molecules 13:2674–2703
Hoang NT, Tóth K, Stacey G (2020) The role of microRNAs in the legume–Rhizobium nitrogen-fixing symbiosis. J Exp Bot 71(5):1668–1680
Hu J, Sun L, Ding Y (2013) Identification of conserved microRNAs and their targets in chickpea (Cicer arietinum L.). Plant Signal Behav 8:e23604
Jerome Jeyakumar JM, Ali A, Wang WM, Thiruvengadam M (2020) Characterizing the role of the miR156-SPL network in plant development and stress response. Plants (Basel) 9:1206
Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI (2018) Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res 46:D335–D342
Khemka N, Singh Rajkumar M, Garg R, Jain M (2021) Genome-wide profiling of miRNAs during seed development reveals their functional relevance in seed size/weight determination in chickpea. Plant Direct 5:e00299
Kim BG, Sung SH, Ahn JH (2012) Biological synthesis of quercetin 3-O-N-acetylglucosamine conjugate using engineered Escherichia coli expressing UGT78D2. Appl Microbiol Biotechnol 93:2447–2453
Kompelli SK, Kompelli VSP, Enjala C, Suravajhala P (2015) Genome-wide identification of miRNAs in pigeonpea (‘Cajanus cajan’L.). Aust J Crop Sci 9:215
Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47:D155–D162
Kumar S, Das M, Sadhukhan A, Sahoo L (2022) Identification of differentially expressed mungbean miRNAs and their targets in response to drought stress by small RNA deep sequencing. Curr Plant Biol 30:100246
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Lazar G, Goodman HM (2006) MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proc Natl Acad Sci USA 103:472–476
Li S (2014) Transcriptional control of flavonoid biosynthesis: fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal Behav 9:e27522
Li Y, Liu X, Cai X, Shan X, Gao R, Yang S, Han T, Wang S, Wang L, Gao X (2017) Dihydroflavonol 4-reductase genes from freesia hybrida Play important and partially overlapping roles in the biosynthesis of flavonoids. Front Plant Sci 8:428
Li C, Zhang B, Chen B, Ji L, Yu H (2018) Site-specific phosphorylation of TRANSPARENT TESTA GLABRA1 mediates carbon partitioning in Arabidopsis seeds. Nat Commun 9:571
Luo X, Gao Z, Shi T, Cheng Z, Zhang Z, Ni Z (2013) Identification of miRNAs and their target genes in peach (Prunus persica L.) using high-throughput sequencing and degradome analysis. PLoS One 8:e79090
Luo Y, Zhang X, Luo Z, Zhang Q, Liu J (2015) Identification and characterization of microRNAs from chinese pollination constant non-astringent persimmon using high-throughput sequencing. BMC Plant Biol 15:11
Mohanty CS, Verma S, Singh V, Khan S, Gaur P, Gupta P, Nizar MA, Dikshit N, Pattanayak R, Shukla A (2013) Characterization of winged bean (Psophocarpus tetragonolobus (L.) DC.) based on molecular, chemical and physiological parameters
Nithin C, Thomas A, Basak J, Bahadur RP (2017) Genome-wide identification of miRNAs and lncRNAs in Cajanus cajan. BMC Genom 18:878
Ó’Maoiléidigh DS, van Driel AD, Singh A, Sang Q, Le Bec N, Vincent C, de Olalla EB, Vayssières A, Romera Branchat M, Severing E, Martinez Gallegos R (2021) Systematic analyses of the MIR172 family members of Arabidopsis define their distinct roles in regulation of APETALA2 during floral transition. PLoS Biology, 19(2), e3001043
Pelaez P, Trejo MS, Iniguez LP, Estrada-Navarrete G, Covarrubias AA, Reyes JL, Sanchez F (2012) Identification and characterization of microRNAs in Phaseolus vulgaris by high-throughput sequencing. BMC Genom 13:83
Pourcel L, Irani NG, Lu Y, Riedl K, Schwartz S, Grotewold E (2010) The formation of anthocyanic vacuolar inclusions in Arabidopsis thaliana and implications for the sequestration of anthocyanin pigments. Mol Plant 3:78–90
Pradhan S, Verma S, Chakraborty A, Bhatia S (2021) Identification and molecular characterization of miRNAs and their target genes associated with seed development through small RNA sequencing in chickpea. Funct Integr Genom 21:283–298
Price ML, Van Scoyoc S, Butler LG (1978) A critical evaluation of the vanillin reaction as an assay for tannin in sorghum grain. J Agric Food Chem 26:1214–1218
Rao MJ, Xu Y, Huang Y, Tang X, Deng X, Xu Q (2019) Ectopic expression of citrus UDP-GLUCOSYL TRANSFERASE gene enhances anthocyanin and proanthocyanidins contents and confers high light tolerance in Arabidopsis. BMC Plant Biol 19:603
Rasool F, Uzair M, Naeem MK, Rehman N, Afroz A, Shah H, Khan MR (2021) Phenylalanine ammonia-lyase (PAL) genes family in wheat (Triticum aestivum L.): genome-wide characterization and expression profiling. Agronomy 11:2511
Rauf A, Imran M, Abu-Izneid T, Iahtisham Ul H, Patel S, Pan X, Naz S, Sanches Silva A, Saeed F, Rasul Suleria HA (2019) Proanthocyanidins: a comprehensive review. Biomed Pharmacother 116:108999
Reddy N, Pierson M, Sathe S, Salunkhe D (1985) Dry bean tannins: a review of nutritional implications. J Am Oil Chem Soc 62:541–549
Roy S, Tripathi AM, Yadav A, Mishra P, Nautiyal CS (2016) Identification and expression analyses of miRNAs from two contrasting flower color cultivars of Canna by deep sequencing. PLoS One 11:e0147499
Rubio-Somoza I, Weigel D (2011) MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16:258–264
Santos-Buelga C, Scalbert A (2000) Proanthocyanidins and tannin‐like compounds–nature, occurrence, dietary intake and effects on nutrition and health. J Sci Food Agric 80:1094–1117
Saslowsky D, Winkel-Shirley B (2001) Localization of flavonoid enzymes in Arabidopsis roots. Plant J 27:37–48
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Sharma D, Tiwari M, Pandey A, Bhatia C, Sharma A, Trivedi PK (2016) MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiol 171:944–959
Shi Y, Xia H, Cheng X, Zhang L (2021) Genome-wide miRNA analysis and integrated network for flavonoid biosynthesis in Osmanthus fragrans. BMC Genom 22:141
Singh V, Goel R, Pande V, Asif MH, Mohanty CS (2017) De novo sequencing and comparative analysis of leaf transcriptomes of diverse condensed tannin-containing lines of underutilized Psophocarpus tetragonolobus (L.) DC. Sci Rep 7:44733
Singh V, Lone RA, Kumar V, Mohanty CS (2023) Reducing the biosynthesis of condensed tannin in winged bean (Psophocarpus tetragonolobus (L.) DC.) by virus-induced gene silencing of anthocyanidin synthase (ANS) gene. 3 Biotech 13(1):16. https://doi.org/10.1007/s13205-022-03435-5
Song C, Wang C, Zhang C, Korir NK, Yu H, Ma Z, Fang J (2010) Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genom 11:431
Sreekumar S, Soniya E (2017) Artificial microRNAs promote high-level production of biomolecules through metabolic engineering of phenylpropanoid pathway. CRC Crit Rev Plant Sci 36:353–366
Sripathi VR, Choi Y, Gossett ZB, Stelly DM, Moss EM, Town CD, Walker LT, Sharma GC, Chan AP (2018) Identification of microRNAs and their targets in four Gossypium species using RNA sequencing. Curr Plant Biol 14:30–40
Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V (2012) The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 28:2059–2061
Sun R, He Q, Zhang B, Wang Q (2015) Selection and validation of reliable reference genes in Gossypium raimondii. Biotechnol Lett 37:1483–1493
Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, Moulton V, Dalmay T (2008) High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genom 9:593
Tan NH, Rahim ZH, Khor HT, Wong KC (1983) Winged bean (Psophocarpus tetragonolobus) tannin level, phytate content, and hemagglutinating activity. J Agric Food Chem 31:916–917
Tan H, Man C, Xie Y, Yan J, Chu J, Huang J (2019) A crucial role of GA-regulated flavonol biosynthesis in root growth of Arabidopsis. Mol Plant 12:521–537
Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939
Thompson EP, Wilkins C, Demidchik V, Davies JM, Glover BJ (2010) An Arabidopsis flavonoid transporter is required for anther dehiscence and pollen development. J Exp Bot 61:439–451
Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45:W122–W129
Tohge T, de Souza LP, Fernie AR (2017) Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J Exp Bot 68:4013–4028
Tuller J, Marquis RJ, Andrade SM, Monteiro AB, Faria LD (2018) Trade-offs between growth, reproduction and defense in response to resource availability manipulations. PLoS One 13:e0201873
Wang B, Xue Y, Zhang Z, Ding D, Fu Z, Tang J (2016) Transcriptomic analysis of maize kernel row number-associated miRNAs between a single segment substitution line and its receptor parent. Plant Growth Regul 78:145–154
Wang Y, Liu W, Wang X, Yang R, Wu Z, Wang H, Wang L, Hu Z, Guo S, Zhang H et al (2020) MiR156 regulates anthocyanin biosynthesis through SPL targets and other microRNAs in poplar. Hortic Res 7:118
Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299:396–399
Xu W, Grain D, Bobet S, Le Gourrierec J, Thévenin J, Kelemen Z, Lepiniec L, Dubos C (2014) Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–b HLH–WDR complexes and their targets in a rabidopsis seed. New Phytol 202:132–144
Xu H, Wang Y, Chen Y, Zhang P, Zhao Y, Huang Y, Wang X, Sheng J (2016) Subcellular localization of galloylated catechins in tea plants [Camellia sinensis (L.) O. Kuntze] assessed via immunohistochemistry. Front Plant Sci 7:728
Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA (2006) Conservation and divergence of plant microRNA genes. Plant J 46:243–259
Zhang X, Gou M, Liu CJ (2013) Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25:4994–5010
Zhao ZC, Hu GB, Hu FC, Wang HC, Yang ZY, Lai B (2012) The UDP glucose: flavonoid-3-O-glucosyltransferase (UFGT) gene regulates anthocyanin biosynthesis in litchi (Litchi chinesis Sonn.) During fruit coloration. Mol Biol Rep 39:6409–6415
Zhao Y, Wei Q, Chen T, Xu L, Liu J, Zhang X, Han G, Ma Q (2022) Identification and characterization of heat-responsive miRNAs and their regulatory network in maize. Plant Growth Regul 96:195–208
Zhu J, Li W, Yang W, Qi L, Han S (2013) Identification of microRNAs in Caragana intermedia by high-throughput sequencing and expression analysis of 12 microRNAs and their targets under salt stress. Plant Cell Rep 32:1339–1349
Zhu Y, Liu Q, Xu W, Yao L, Wang X, Wang H, Xu Y, Li L, Duan C, Yi Z et al (2021) Identification of novel drought-responsive miRNA regulatory network of drought stress response in common vetch (Vicia sativa). Open Life Sci 16:1111–1121
Acknowledgements
CSM and SPN acknowledge the Department of Biotechnology for the financial support to carry out this activity and Director, CSIR-National Botanical Research Institute for providing the infrastructure facility. We also acknowledge the National Bureau of Plant Genetic Resources for providing the required germplasm. PP and SPN acknowledge the University Grants Commission for a research fellowship grant. The authors also acknowledge Kirti Pandey and Arpit Chauhan for providing experimental support during the study.
Funding
The research was funded by the project “Bioresources and sustainable livelihoods of Northeast India”, Department of Biotechnology, Govt. of India; Pro.
Author information
Authors and Affiliations
Contributions
CSM, SPN, and VS designed the research work. SPN conducted most of the experiments and prepared samples for small RNA sequencing. PP performed all bioinformatics analyses. SPN and PP wrote the manuscript. VS and AMT provided the necessary suggestions for conducting experiments. CSM, VS, and SB helped in finalising the manuscript and critically assessing the report. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest
Additional information
Communicated by Mohsin Tanveer.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10725_2023_971_MOESM1_ESM.tif
Supplementary material 1 (TIF 9236.5 kb)—Fig.S1 HPLC Chromatogram of standards (catechin, epicatechin and epigallocatechin)and methanolic (MeOH) extract of P. tetragonolobus leaves
10725_2023_971_MOESM2_ESM.tif
Supplementary material 2 (TIF 12163.6 kb)—Fig.S2 Nucleotide length variations of small RNAs in the sequenced library of HPW and LPW in P. tetragonolobus
10725_2023_971_MOESM3_ESM.tif
Supplementary material 3 (TIF 6538.9 kb)—Fig.S3 Correlation plot between the replicate 1 and replicate 2 of sequenced HPW and LPW library. R value represents correlation value of 0.93 and0.82 in HPW and LPW respectively with significant p-value < 2.2e-16
10725_2023_971_MOESM4_ESM.tif
Supplementary material 4 (TIF 7247.2 kb)—Fig.S4 Different number of identified conserved miRNAs in HPW and LPW library in 52 plant lineages. X-axis represented the different plant species while different numbers of miRNAs was scaled in Y-axis. Bold font scientific name was used for plant species that belongs to Poaceae family
10725_2023_971_MOESM5_ESM.tif
Supplementary material 5 (TIF 9050.7 kb)—Fig.S5 Volcano plot to represents the differentially expressed miRNAs between the HPW and LPW lines of P. tetragonolobus. X-axis was used for the log2 Fold change whereas Y-axis representing the –log2 (p-value). Each dot describes the expressed miRNAs; wherein red dots define the differentially expressed miRNAs
10725_2023_971_MOESM6_ESM.tif
Supplementary material 6 (TIF 10990.4 kb)—Fig.S6 Heat map visualizations of log2 fold change expression values of miRNAs fromthe HPW and LPW. Lower expression values are represented in the red color whilehigher expression values are illustrated in green color. The color scale isranged from 1 to 14
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nayak, S.P., Prasad, P., Singh, V. et al. Role of miRNAs in the regulation of proanthocyanidin biosynthesis in the legume Psophocarpus tetragonolobus (L.) DC.. Plant Growth Regul 102, 23–38 (2024). https://doi.org/10.1007/s10725-023-00971-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-023-00971-9