Abstract
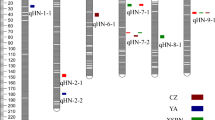
A genome-wide association Study (GWAS) detects linkages between markers and traits of interest for application in genomics-guided breeding. This study utilized GWAS to identify quantitative trait loci (QTL) associated with important agronomic traits in selected palms from Malaysian Palm Oil Board’s (MPOB) African germplasm collection. A total of 635 African oil palm accessions from Angola, Madagascar, Nigeria, and Tanzania were genotyped using a 4451 Single Nucleotide Polymorphism (SNP) Chip via the Illumina platform. Subsequently, genetic diversity was analyzed using 1464 informative SNP markers, and the Angola germplasm was discovered to have the highest heterozygosity and, as expected, Madagascar the lowest. Population structure analysis revealed two main subpopulations, Madagascar versus the others (Angola, Nigeria, and Tanzania). Linkage disequilibrium (LD) analysis conducted using Haploview 4.2 suggested LD decay to be 1.9 kb at r2 = 0.1. Based on fixed and random model calculating probability unification (FarmCPU) analyses, 49 markers were significantly associated with 17 phenotypic traits, some located within the genomic regions associated with similar traits in other studies. QTL on chromosomes 6, 9, and 11 are common among the germplasm and advanced populations, thereby suggesting that the genomic region is selectively fixed in advanced breeding lines. This adds confidence to their potential utility in breeding programs to enhance selection efficiency and progress of the genetic improvement of a perennial crop like oil palm. Interestingly, protein STRUBBELIG-RECEPTOR family 6 and Beta-galactosidase 11 genes were also observed in the QTL intervals, which are potential candidates in expression studies to further dissect the genetic architecture of the traits.











Similar content being viewed by others
Data availability
The datasets generated from this study are available from the corresponding author upon reasonable request.
References
Abecasis GR, Noguchi E, Heinzmann A, Traherne JA, Bhattacharyya S, Leaves NI, Anderson GG, Zhang Y, Lench NJ, Carey A, Cardon LR, Moffat MF, Cookson WOC (2001) Extent and distribution of linkage disequilibrium in three genomic regions. Am J Genet 68:191–197. https://doi.org/10.1086/316944
Ameen M, Zamri NM, May ST, May ST, Azizan MT, Aqsha A, Sabzoi N, Sher F (2022) Effect of acid catalysts on hydrothermal carbonization of Malaysian oil palm residues (leaves, fronds, and shells) for hydrochar production. Biomass Conv Bioref 12:103–114. https://doi.org/10.1007/s13399-020-01201-2
Arif IA, Bakir MA, Khan HA, Ahamed A, Al Farhan AH, Al Homaidan AA, Sadoon MA, Bahkali AH, Shobrak M (2010) A simple method for DNA extraction from mature date palm leaves: impact of sand grinding and composition of lysis buffer. Int J Mol Sci 11(9):3149–3157. https://doi.org/10.3390/ijms11093149
Babu BK, Mathur RK, Anitha P, Ravichandran G, Bhagya HP (2021) Phenomics, genomics of oil palm (Elaeis guineensis Jacq.): way forward for making sustainable and high yielding quality oil palm. Physiol Mol Biol Plants 27(3):587–604. https://doi.org/10.1007/s12298-021-00964-w
Babu BK, Mathur RK, Ravichandran G, Anita P, Venu MVB (2020) Genome wide association study (GWAS) and identification of candidate genes for yield and oil yield related traits in oil palm (Elaeis guineensis) using SNPs by genotyping-based sequencing. Genomics 112(1):1011–1020. https://doi.org/10.1016/j.ygeno.2019.06.018
Babu BK, Mathur RK, Ravichandran G, Venu MVB (2019) Genome-wide association study (GWAS) for stem height increment in oil palm (Elaeis guineensis) germplasm using SNP markers. Tree Genetics Genomes 15:3. https://doi.org/10.1007/s11295-019-1349-2
Bakoumé C, Wickneswari R, Siju S, Rajanaidu N, Kushairi A, Billotte N (2015) Genetic diversity of the world’s largest oil palm (Elaeis guineensis Jacq) field genebank accessions using microsatellite markers. Genet Resour Crop Evol 62(3):349–360. https://doi.org/10.1007/s10722-014-0156-8
Balasubramaniam S, Lee HC, Lazan H, Othman R, Ali ZM (2005) Purification and properties of a beta-galactosidase from carambola fruit with significant activity towards cell wall polysaccharides. Phytochemistry 66(2):153–163. https://doi.org/10.1016/j.phytochem.2004.11.005
Barcelos E, Rios SA, Cunha RNV, Lopes R, Motoike SY, Babiychuk E, Skirycz A, Kushnir S (2015) Oil palm natural diversity and the potential for yield improvement. Front Plant Sci 6:1–16. https://doi.org/10.3389/fpls.2015.00190
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265. https://doi.org/10.1093/bioinformatics/bth457
Biles CL, Bruton BD, Russo V, Wall MM (1997) Characterisation of β-Galactosidase Isozymes of ripening peppers. J Sci Food Agric 75:237–243
Billote N, Jourjon MF, Marseillac N, Berger A, Flori A, Asmady H, Adon B, Sing R, Nouy B, Potier F, Cheah SC, Rohde W, Ritter E, Courtois B, Charrier A, Mangin B (2010) QTL detection by multi-parent linkage mapping in oil palm (Elaeis guineensis Jacq.). Theor Appl Genet 120:1673–1687
Chamarthi SK, Kaler AS, Abdel-Haleem H, Fritschi FB, Gillman JD, Ray JD, Smith JR, Dhanapal AP, King CA, Purcell LC (2021) Identification and confirmation of loci associated with canopy wilting in soybean using genome-wide association mapping. Front Plant Sci 12:698116. https://doi.org/10.3389/fpls.2021.698116
Chantarangsee M, Tanthanuch W, Fujimura T, Fry SC, Cairns JK (2007) Molecular characterization of β-galactosidase from germinating rice (Oryza sativa). Plant Sci 173:118–134
Chaudhary A, Chen X, Gao J, Leśniewska B, Hammerl R, Dawid C, Schneitz K (2020) The Arabidopsis receptor kinase STRUBBELIG regulates the response to cellulose deficiency. PLoS Genet 16(1):e1008433. https://doi.org/10.1371/journal.pgen.1008433
Chen SY, Lai MH, Tung CW, Wu DH, Chang FY, Lin TC, Chung CL (2019) Genome-wide association mapping of gene loci affecting disease resistance in the rice-Fusarium fujikuroi pathosystem. Rice 12(1):1–2
Chevalier D, Batoux M, Fulton L, Pfister K, Yadav RK, Schellenberg M, Schneitz K (2005) STRUBBELIG defines a receptor kinase-mediated signalling pathway regulating organ development in Arabidopsis. Proc Natl Acad Sci U S A 102(25):9074–9079. https://doi.org/10.1073/pnas.0503526102
Corley RHV, Tinker PB (2016) The oil palm, 5th edn. Wiley Blackwell
Dean GH, Zheng H, Tewari J, Huang J, Young DS, Hwang YT, Western TL, Carpita NC, McCann MC, Mansfield SD, Haughn GW (2007) The Arabidopsis MUM2 gene encodes a β-Galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19(12):4007–4021
Dresselhaus T, Srilunchang KO, Leljak-Levanić D, Schreiber DN, Garg P (2006) The fertilization-induced DNA replication factor MCM6 of maize shuttles between cytoplasm and nucleus, and is essential for plant growth and development. Plant Physiol 140(2):512–527. https://doi.org/10.1104/pp.105.074294
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genet Resour 4(2):359–361. https://doi.org/10.1007/s12686-011-9548-7
Esteban R, Dopico B, Muñoz FJ, Romo S, Martín I, Labrador E (2003) Cloning of a Cicer arietinum β-galactosidase with pectin-degrading function. Plant Cell Physiol 44(7):718–725. https://doi.org/10.1093/pcp/pcg087
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol 14(8):2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Eyüboglu B, Pfister K, Haberer G, Chevalier D, Fuchs A, Mayer KFX Schneitz K (2007) Molecular characterization of the STRUBBELIG-RECEPTOR FAMILY of genes encoding putative leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. BMC Plant Biol 7(16).
Fadila AM, Norziha A, Din MA, Rajanaidu N, Kushairi A (2016) Evaluation of bunch index in MPOB oil palm (Elaeis guineensis Jacq) germplasm collection. J Oil Palm Res 28(4):442–451
Figueiredo SA, Lashermes P, Aragão FJL (2011) Molecular characterization and functional analysis of the β-galactosidase gene during Coffea arabica (L.) fruit development. J Exp Bot 62(8):2691–2703. https://doi.org/10.1093/jxb/erq440
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54(1):357–374. https://doi.org/10.1146/annurev.arplant.54.031902.134907
Fragoso CA, Moreno M, Wang Z, Heffelfinger C, Arbelaez LJ, Aguirre JA, Franco N, Romero LE, Labadie K, Zhao H, Dellaporta SL (2017) Genetic architecture of a rice nested association mapping population. G3 7(6):1913–1926
Gupta PK, Kulwal PL, Jaiswal V (2014) Association mapping in crop plants. In: Advances in genetics, vol 85. Elsevier, pp 109–147
Glémin S, François CM, Galtier N (2019) Genome evolution in outcrossing vs. selfing vs. asexual species. In: Anisimova M. (ed) Evolutionary genomics. Methods in molecular biology, vol 1910. Humana, New York. https://doi.org/10.1007/978-1-4939-9074-0_11
Hill WG, Weir BS (1988) Variances and covariances of squared linkage disequilibria in finite populations. Theor Popul Biol 33(1):54–78. https://doi.org/10.1016/0040-5809(88)90004-4
Höglund J, Rafati N, Rask-Andersen M, Enroth S, Karlsson T, Ek WE, Johansson Å (2019) Improved power and precision with whole genome sequencing data in genome-wide association studies of inflammatory biomarkers. Sci Rep 6:16844. https://doi.org/10.1038/s41598-019-53111-7
Huang X, Han B (2013) Natural variations and genome-wide association studies in crop plants. Annu Rev Plant Biol 65(1):531–551. https://doi.org/10.1146/annurev-arplant-050213-035715
Hou F, Du T, Qin Z, Xu T, Li A, Dong S, Ma D, Li Z, Wang Q, Zhang L (2021) Genome-wide in silico identification and expression analysis of beta-galactosidase family members in sweet potato [Ipomoea batatas (L) Lam]. BMC Genomics 22:140. https://doi.org/10.1186/s12864-021-07436-1
Ithnin M, Rajanaidu N, Zakri AH, Cheah SC (2006) Assessment of genetic diversity in oil palm (Elaeis guineensis Jacq) using restriction fragment length polymorphism (RFLP). Genet Resour Crop Evol 53(1):187–195. https://doi.org/10.1007/s10722-004-4004-0
Ithnin M, Xu Y, Marjuni M, Serdari NM, Amiruddin MD, Low E-TL, Tan Y-C, Yap S-J, Ooi LCL, Nookiah R, Singh R, Xu S (2017) Multiple locus genome-wide association studies for important economic traits of oil palm. Tree Genet Genomes 13:5. https://doi.org/10.1007/s11295-017-1185-1
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-Box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143(4):1467–1483. https://doi.org/10.1104/pp.106.091900
Jeennor S, Volkaert H (2014) Mapping of quantitative trait loci (QTLs) for oil yield using SSRs and gene-based markers in African oil palm (Elaeis guineensis Jacq.). Tree Genet Genomes 10:1–14
Jin J, Lee M, Bai B, Sun Y, Qu J, Rahmadsyah AY, Lim CH, Suwanto A, Sugiharti M, Wong L, Ye J, Chua NH, Yue GH (2016) Draft genome sequence of an elite Dura palm and whole-genome patterns of DNA variation in oil palm. DNA Res 23(6):527–533. https://doi.org/10.1093/dnares/dsw036
John Martin JJ, Yarra R, Wei L, Cao H (2022) Oil palm breeding in the modern era: challenges and opportunities. Plants. https://doi.org/10.3390/plants11111395
Kaler AS, Ray JD, Schapaugh WT, King CA, Purcell LC (2017) Genome-wide association mapping of canopy wilting in diverse soybean genotypes. Theor Appl Genet 130:2203–2217. https://doi.org/10.1007/s00122-017-2951-z
Kim M-S, Ko S-R, Le VT, Jee M-G, Jung YJ, Kang K-K, Cho Y-G (2022) Development of SNP markers from GWAS for selecting seed coat and aleurone layers in brown rice (Oryza sativa L.). Genes 13:1805. https://doi.org/10.3390/genes13101805
Kumar J, Agrawal V (2019) Assessment of genetic diversity, population structure and sex identification in dioecious crop, Trichosanthes dioica employing ISSR, SCoT and SRAP markers. Heliyon 5:e01346. https://doi.org/10.1016/j.heliyon.2019.e01346
Kwong QB, Teh CK, Ong AL, Heng HY, Lee HL, Mohamed M, Low JZ, Apparow S, Chew FT, Mayes S, Kulaveerasingam H, Tammi M, Appleton DR (2016) Development and validation of a high-density SNP genotyping array for African oil palm. Mol Plant 9(8):1132–1141. https://doi.org/10.1016/j.molp.2016.04.010
Lally RD, Donaleshen K, Chirwa U, Eastridge K, Saintilnord W, Dickinson E, Murphy R, Borst S, Horgan K, Dawson K (2021) Transcriptomic response of huanglongbing-infected citrus sinensis following field application of a microbial fermentation product. Front Plant Sci 12:754391. https://doi.org/10.3389/fpls.2021.754391
Lechner E, Archard P, Vansiri A, Potuschak T, Genschik P (2006) F-box proteins everywhere. Curr Opin Plant Biol 9(6):631–638. https://doi.org/10.1016/j.pbi.2006.09.003
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28(18):2397–2399. https://doi.org/10.1093/bioinformatics/bts444
Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLOS Genet 12(2):e1005767. https://doi.org/10.1371/journal.pgen.1005767
Liu S, Liu X, Zhang X, Chang S, Ma C, Qin F (2022) Co-expression of ZmVPP1 with ZmNAC111 confers robust drought resistance in maize. Genes 14(1):8. https://doi.org/10.3390/genes14010008
Longmei N, Gill GK, Zaidi PH, Kumar R, Nair SK, Hindu V, Vinayan MT, Vikal Y (2021) Genome wide association mapping for heat tolerance in sub-tropical maize. BMC Genomics 22(1):1–4
Low ETL, Halim MAA, Rosli R, Chan KL, Azizi N, Nagappan J, Sanusi NSNM, Amiruddin N, Razak ASA, Kadri F, Singh R, Ong MA, Sambanthamurthi R. (2015) MYPalmViewer: oil palm genome browser. MPOB Information Series No 148. ISSN 1511–7871. http://palmoilis.mpob.gov.my/TOTV3/wp-content/uploads/2020/02/TS148.pdf. Accessed 13 September 2023.
Mather KA, Caidedo AL, Polato NR, Olsen KM, McCouch S, Purugganan MD (2007) The extent of linkage disequilibrium in Rice (Oryza sativa L.) genetics 177(4): 2223–2232. https://doi.org/10.1534/genetics.107.079616
Marroni F, Pinosio S, Zaina G, Fogolari F, Felice N, Cattonaro F, Morgante M (2011) Nicleotide diversity and linkage disequilibrium in Populus nigra cinnamyl alcohol dehydrogenase (CAD4) gene. Tree Genetics Genome 7:1011–1023
Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, Buckler ES (2009) Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21(8):2194–2202
Oil World (2021). Oil world monthly. No 50, vol 64. 17 December 2021. ISTA Mielke GmbH, Germany.
Ong PW, Maizura I, Marhalil M, Rajanaidu N, Abdullah NAP, Rafii MY, Ooi LCL, Low ETL, Singh R (2018) Association of SNP markers with height increment in MPOB-Angolan natural oil palm populations. J Oil Palm Res 30(1):61–70. https://doi.org/10.21894/jopr.2017.0003
Ooi LCL, Low ETL, Ong-Abdullah M, Rajanaidu N, Ting NC, Nagappan J, Manaf MAA, Chan KL, Halim MA, Azizi N, Omar W, Murad AJ, Lakey N, Ordway JM, Favello A, Budiman MA, Brunt AV, Beil M, Leininger MT, Jiang N, Smith SW, Brown CR, Kuek ACS, Bahrain S, Hoynes-O’Connor A, Nguyen AY, Chaudhari HG, Shah SA, Choo YM, Sambanthamurthi R, Singh R (2016) Non-tenera contamination and the economic impact of SHELL genetic testing in the Malaysian independent oil palm industry. Front Plant Sci. https://doi.org/10.3389/fpls.2016.00771
Pan H, Sun Y, Qiao M, Qi H (2022) Beta-galactosidase gene family genome-wide identification and expression analysis of members related to fruit softening in melon (Cucumis melo L.). BMC Genomics 23:1. https://doi.org/10.1186/s12864-022-09006-5
Parveez GKA, Kamil NN, Zawawi NZ, Ong-Abdullah M, Rasuddin R, Soh KL, Selvaduray KR, Seng SH, Idris Z (2022) Oil palm economic performance in Malaysia and R&D progress in 2021. J Oil Palm Res 34(2):185–218. https://doi.org/10.21894/jopr.2022.0036
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Peakall R, Smouse PE (2012) GenAlex 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28(19):2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Perrier X, Jacquemoud-Collet JP (2006) DARwin Software. http://darwin.cirad.fr/
Prasanna V, Prabha TN, Tharanathan RN (2005) Multiple forms of β-galactosidase from mango (Mangifera indica L Alphonso) fruit pulp. J Sci Food Agric 85:797–803
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Rahimah AR, Cheah SC, Rajinder S (2006) Freeze-drying of oil palm (Elaeis guineensis) leaf and its effect on the quality of extractable DNA. J Oil Palm Res 18:296–304
Rahman M, Hoque A, Roy J (2022) Linkage disequilibrium and population structure in a core collection of Brassica napus (L.). PLoS ONE 17(3):e0250310. https://doi.org/10.1371/journal.pone.0250310
Rajanaidu N, Kushairi A, Amiruddin MD (2017) Oil palm genetic resources exploration. In: Monograph oil palm genetic resources. Perpustakaan Negara Malaysia, pp 37–91
Rajora OP, Fageria MS, Fitzsimmons M (2023) Forest fires in genetic diversity and population structure of boreal conifer, white spruce (Picea glauca (Moench) Voss): Implications for genetic resource management and adaptive potential under climate change. Forests. https://doi.org/10.3390/f14010157
Rance KA, Mayes S, Price Z, Jack PL, Corley RHV (2001) Quantitative trait loci for yield components in oil palm (Elaeis guineensis Jacq). Theor Appl Genet 103(8):1302–1310. https://doi.org/10.1007/s122-001-8204-z
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebly J, Kresovich S, Goodman MM, Buckler ES (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. PNAS 98(20):11479–11484
Ross GS, Wegrzyn T, MacRae EA, Redgwell RJ (1994) Apple β-Galactosidase. Activity against cell wall polysaccharides and characterization of a related cDNA clone. Plant Physiol 106:521–528
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Sehgal D, Rosyara U, Mondal S, Singh R, Poland J, Dreisigacker S (2020) Incorporating genome-wide association mapping result into genomic prediction models for grain yield and yield stability in CIMMYT spring bread wheat. Front Plant Sci. https://doi.org/10.3389/fpls.2020.00197
Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. PNAS 98(19):10763–10768
Shultz RW, Lee TJ, Allen GC, Thompson WF, Hanley-Bowdoin L (2009) Dynamic localization of the DNA replication proteins MCM5 and MCM7 in plants. Plant Physiol 150(2):658–669. https://doi.org/10.1104/pp.109.136614
Singh R, Low ETL, Ooi LCL, Ong-Abdullah M, Ting NC, Nagappan J, Rajanaidu N, Amiruddin MD, Rosli R, Manaf MAA, Chan KL, Halim MA, Azizi N, Lakey N, Smith SW, Budiman MA, Hogan M, Bacher B, Brunt AV, Wang C, Ordway JM, Sambanthamurthi R, Martienssen RA (2013a) The oil palm SHELL gene controls oil yield and encodes a homologue of SEEDSTICK. Nature 500(7462):340–344. https://doi.org/10.1038/nature12356
Singh R, Ong-Abdullah M, Low ETL, Manaf MAA, Rosli R, Nookiah R, Ooi LCL, Ooi SE, Chan KL, Halim MA, Azizi N, Nagappan J, Bacher B, Lakey N, Smith SW, He D, Hogan M, Budiman MA, Lee EK, DeSalle R, Kudrna D, Goicoechea JL, Wing RA, Wilson RK, Fulton RS, Ordway JM, Martienssen RA, Sambanthamurthi R (2013b) Oil palm genome sequence reveals divergence of interfertile species in old and new worlds. Nature 500(7462):335–339. https://doi.org/10.1038/nature12309
Singh R, Low ETL, Ooi LCL, Ong-Abdullah M, Rajanaidu N, Ting NC, Marjuni M, Chan PL, Ithnin M, Manaf MAA, Nagappan J, Chan KL, Rosli R, Halim MA, Azizi N, Budiman MA, Lakey N, Bacher B, Brunt AV, Wang C, Hogan M, He D, MacDonald JD, Smith SW, Ordway JM, Martienssen RA, Sambanthamurthi R (2014) The oil palm VIRESCENS gene control fruit color and encodes a R2R3-MYB. Nat Commun. https://doi.org/10.1038/ncomms5106
Singh R, Kamil NN, Ooi LCL, Low ETL, Abdullah MO, Sambanthamurthi R, Lakey N, Manaf MAA, Ismail A (2022) SHELL genetic testing: a key enabler of yield improvement and sustainable palm oil production. The Planter 98(1159):713–727. https://doi.org/10.56333/tp.2022.011
Smith DL, Gross KC (2000) A family of at least seven β-galactosidase genes is expressed during tomato fruit development. Plant Physiol 123(3):1173–1183. https://doi.org/10.1104/pp.123.3.1173
Soh AC, Mayes S, Roberts J, Rajanaidu N, Amiruddin MD, Marhalil M, Norziha A, Ong-Abdullah M, Fadila AM, Azwani ABN, Anak LA, Zulkifli Y, Mustafa S, Ithnin M, Kushairi A, Barcelos E, Amblard P (2017) Genetic resources. In: Soh AC, Mayes S, Roberts JA (eds) Oil palm breeding: genetics and genomic, 1st edn. CRC Press, New York
Spindel J, Begum H, Akdemir D, Collard B, Redoña E, Jannink J-L, McCouch S (2016) Genome-wide prediction models that incorporate de novo GWAS are a powerful new tool for tropical rice improvement. Heredity 116:395–408. https://doi.org/10.1038/hdy.2015.113
Steketee CJ, Schapaugh WT, Carter TE Jr, Li Z (2020) Genome-wide association analyses reveal genomic regions controlling canopy wilting in soybean. G3 (bethesda) 10(4):1413–1425. https://doi.org/10.1534/g3.119.401016
Sun C, Wang B, Wang X, Hu K, Li K, Li Z, Li S, Yan L, Guan C, Zhang J, Zhang Z, Chen S, Wen J, Tu J, Shen J, Fu T, Yi B (2016) Genome-wide association study dissecting the genetic architecture underlying the branch angle trait in rapeseed (Brassica napus L). Sci Rep. https://doi.org/10.1038/srep33673
Suzana M, Rahimah AR, Maizura I, Singh R (2015) A simple and rapid protocol for isolation of genomic DNA from oil palm leaf tissue. J Oil Palm Res 27(3):282–287
Swaray S, Din Amiruddin M, Rafii MY, Jamian S, Ismail MF, Jalloh M, Marjuni M, Mustakim Mohamad M, Yusuff O (2020) Influence of parental dura and pisifera genetic origins on oil palm fruit set ratio and yield components in their d × p progenies. Agronomy 10(11):1793. https://doi.org/10.3390/agronomy10111793
Tateishi A, Inoue H, Shiba H, Yamaki S (2001) Molecular cloning of β-Galactosidase from Japanese Pear (Pyrus pyrifolia) and its gene expression with fruit ripening. Plant Cell Physiol 42(5):492–498
Teh CK, Ong AL, Kwong QB, Apparow S, Chew FT, Mayes S, Mohamed M, Appleton D, Kulaveerasingam H (2016) Genome-wide association study identifies three key loci for high mesocarp oil content in perennial crop oil palm. Sci Rep 6:1–7. https://doi.org/10.1038/srep19075
Thomson AL, Moreau RE (1957) Feeding habits of the palm-nut vulture Gypohierax. Ibis 99(4):608–613
Ting NC, Jansen J, Mayes S, Massawe F, Sambanthamurthi R, Ooi LCL, Chin CW, Arulandoo X, Seng TY, Alwee SSRS, Ithnin M, Singh R (2014) High density SNP and SSR-based genetic maps of two independent oil palm hybrids. BMC Genomics 15(1):309. https://doi.org/10.1186/1471-2164-15-309
Ting NC, Yaakub Z, Kamaruddin K, Mayes S, Massawe F, Sambanthamurthi R, Jansen J, Low ETL, Ithnin M, Kushairi A, Arulandoo X, Rosli R, Chan KL, Amiruddin N, Sritharan K, Lim CC, Rajanaidu N, Amiruddin MD, Singh R (2016) Fine-mapping and cross-validation of QTL linked to fatty acid composition in multiple independent interspecific crosses of oil palm. BMC Genomics. https://doi.org/10.1186/s12864-016-2607-4
Trainotti L, Spinello R, Piovan A, Spolaore S, Casadoro G (2001) β-Galactosidase with a lectin-like domain are expressed in strawberry. J Exp Bot 52(361):1635–1645
Tranbarger TJ, Fooyontphanich K, Roongsattham P, Pizot M, Collin M, Jantasuriyarat C, Suraninpong P, Tragoonrung S, Dussert S, Verdeil JL, Morcillo F (2017) Transcriptome analysis of cell wall and NAC domain transcription factor genes during Elaeis guineensis fruit ripening: evidence for widespread conservation within monocot and eudicot lineages. Front Plant Sci 8:603. https://doi.org/10.3389/fpls.2017.00603
Turuspekov Y, Baibulatova A, Yermekbayev K, Tokhetova L, Chudinov V, Sereda G, Ganal M, Griffiths S, Abugalieva S (2017) GWAS for plant growth stages and yield components in spring wheat (Triticum aestivum L.) harvested in three regions of Kazakhstan. BMC Plant Biol 17(1):1–11
Tuteja N, Tran NQ, Dang HQ, Tuteja R (2011) Plant MCM proteins: role in DNA replication and beyond. Plant Mol Biol 77(6):537–545. https://doi.org/10.1007/s11103-011-9836-3
Vaddepalli P, Fulton L, Batoux M, Yadav RK, Schneitz K (2011) Structure-function analysis of strubbelig, an Arabidopsis atypical receptor-like kinase involved in tissue morphogenesis. PLoS ONE 6:5. https://doi.org/10.1371/journal.pone.0019730
Vanderzande S, Micheletti D, Troggio M, Davey MW, Keulemans J (2017) Genetic diversity, population structure, and linkage disequilibrium of elite and local apple accessions from Belgium using the IRSC array. Tree Genetics Genomes 13:125. https://doi.org/10.1007/s11295-017-1206-0
Vanous A, Gardner C, Blanco M, Martin-Schwarze A, Lipka AE, Flint-Garcia S, Bohn M, Edwards J, Lübberstedt T (2018) Association mapping of flowering and height traits in germplasm enhancement of maize doubled haploid (GEM-DH) lines. Plant Genome 11(2):170083
Wang X, Wang H, Liu S, Ferjani A, Li J, Yan J, Yang X, Qin F (2016) Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat Genet 48(10):1233–1241. https://doi.org/10.1038/ng.3636
Wang H, Chen Y, Wu X, Long Z, Sun C, Wang H, Wang S, Birch PRJ, Tian Z (2018) A potato STRUBBELIG-RECEPTOR FAMILY member, StLRPK1, associates with StSERK3A/BAK1 and activates immunity. J Exp Bot 69(22):5573–5586. https://doi.org/10.1093/jxb/ery310
Wei T, Simko V (2021) R package ‘corrplot’: visualization of a correlation matrix (version 0.92). https://github.com/taiyun/corrplot
Wu Z, Burns JK (2004) A β-galactosidase gene is expressed during mature fruit abscission of ‘Valencia’ orange (Citrus sinensis). J Exp Bot 55(402):1483–1490. https://doi.org/10.1093/jxb/erh163
Xia W, Luo T, Zhang W, Mason AS, Huang D, Huang X, Tang W, Dou Y, Zhang C, Xiao Y (2019) Development of high-density SNP markers and their application in evaluating genetic diversity and population structure in Elaeis guineensis. Front Plant Sci 10:130. https://doi.org/10.3389/fpls.2019.00130
Xiao Y, Liu H, Wu L, Warburton M, Yan J (2017) Genome-wide association studies in maize: praise and stargaze. Mol Plant 10:359–374. https://doi.org/10.1016/j.molp.2016.12.008
Xu G, Ma H, Nei M, Kong H (2008) Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. PNAS 106(3):835–840
Yadav RK, Fulton L, Batoux M, Schneitz K (2008) The Arabidopsis receptor-like kinase STRUBBELIG mediates inter-cell-layer signaling during floral development. Dev Biol 323(2):261–270. https://doi.org/10.1016/j.ydbio.2008.08.010
Yan J, Shah T, Warburton ML, Buckler ES, McMullen MD, Crouch J (2009) Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS ONE 4(12):8451. https://doi.org/10.1371/journal.pone.0008451
Zegeye H, Rasheed A, Makdis F, Badebo A, Ogbonnaya FC (2014) Genome-wide association mapping for seedling and adult plant resistance to stripe rust in synthetic hexaploid wheat. PLoS ONE 9(8):105593
Zeven AC (1967) The semi-wild oil palm and its industry in Africa. Centre for Agricultural Publications and Documentation.
Zeven AC (1972) The partial and complete domestication of the oil palm (Elaeis guineensis). Econ Bot 26(3):274–279
Zhang X, Gonzalez-Carranza ZH, Zhang S, Miao Y, Liu C-J, Roberts JA (2019) F-box proteins in plants. Annu Plant Rev 2:307–328. https://doi.org/10.1016/j.cell.2009.08.031
Zheng M, Peng C, Liu H, Tang M, Yang H, Li X, Liu J, Sun X, Wang X, Hua W, Wang H (2017) Genome-wide association study reveals candidate genes for control of plant height, branch initiation height and branch number in rapeseed (Brassica napus L). Front Plant Sci 8:1246. https://doi.org/10.3389/fpls.2017.01246
Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L, Zhao Y, Ma Y, Fang C, Shen Y, Liu T, Li C, Li Q, Wu M, Wang M, Wu Y, Dong Y, Wan W, Wang X, Ding Z, Gao Y, Xiang H, Zhu B, Lee S-H, Wang W, Tian Z (2015) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 33:408–414. https://doi.org/10.1038/nbt.3096
Zhu XM, Shao XY, Pei YH, Guo XM, Li J, Song XY, Zhao MA (2018) Genetic diversity and genome-wide association study of major ear quantitative traits using high-density SNPs in maize. Front Plant Sci 9:966. https://doi.org/10.3389/fpls.2018.00966
Zolkafli SH, Ting NC, Sanusi NSNM, Ithnin M, Mayes S, Massawe F, Sambanthamurthi R, Ismail I, Abidin MIZ, Roowi SH, Lee YP, Hanafi NFF, Singh R (2021) Comparison of quantitative trait loci (QTL) associated with yield components in two commercial Dura x Pisifera breeding crosses. Euphytica 217:104
Zulkifli Y, Maizura I, Rajinder S (2012) Evaluation of MPOB oil palm germplasm (Elaeis guineensis) populations using EST-SSR. J Oil Palm Res 24:1368–1377
Acknowledgements
The authors would like to thank the Director-General of MPOB for permission to publish this paper. We also would like to express our appreciation to all the members of the Genomics unit, Breeding and Tissue Culture unit, and Bioinformatics unit for their technical support.
Funding
This study was fully funded by Malaysian Palm Oil Board (MPOB).
Author information
Authors and Affiliations
Contributions
MI and RS conceived and design the study. NMS prepared the samples, analyzed the data, and wrote the manuscript. NSNMS provides the genetic map data. SM provides the phenotype data. JS, IM, RS, MMR, and MI reviewed and edited the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that there are no competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Serdari, N.M., Sanusi, N.S.N.M., Suzana, M. et al. Association mapping of selected oil palm germplasm reveals novel and known genomic regions influencing vegetative and bunch component traits. Genet Resour Crop Evol 71, 1709–1735 (2024). https://doi.org/10.1007/s10722-023-01723-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-023-01723-4