Abstract
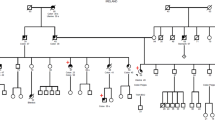
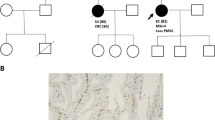
Germline pathogenic variants in the DNA mismatch repair genes (MMR): MLH1, MSH2, MSH6, and PMS2, are causative of Lynch syndrome (LS). However, many of the variants mapping outside the invariant splice site positions (IVS ± 1, IVS ± 2) are classified as variants of unknown significance (VUS). Three such variants (MLH1 c.588+5G>C, c.588+5G>T and c.677+5G>A) were identified in 8 unrelated LS families from Argentina, Brazil and Chile. Herein, we collected clinical information on these families and performed segregation analysis and RNA splicing studies to assess the implication of these VUS in LS etiology. Pedigrees showed a clear pattern of variant co-segregation with colorectal cancer and/or other LS-associated malignancies. Tumors presented deficient expression of MLH1-PMS2 proteins in 7/7 of the LS families, and MSI-high status in 3/3 cases. Moreover, RNA analyses revealed that c.588+5G>C and c.588+5G>T induce skipping of exon 7 whereas c.677+5G>A causes skipping of exon 8. In sum, we report that the combined clinical findings in the families and the molecular studies provided the evidences needed to demonstrate that the three MLH1 variants are causative of LS and to classify c.588+5G>C and c.677+5G>A as class 5 (pathogenic), and c.588+5G>T as class 4 (likely-pathogenic). Our findings underline the importance of performing clinical and family analyses, as well as RNA splicing assays in order to determine the clinical significance of intronic variants, and contribute to the genetic counseling and clinical management of patients and their relatives.


Similar content being viewed by others
Availability of data and material statement
Data from the participating hereditary cancer registers, this is indeed available for researchers following direct contact with the register (thus not freely available online).
References
Carethers JM, Stoffel EM (2015) Lynch syndrome and Lynch syndrome mimics: the growing complex landscape of hereditary colon cancer. World J Gastroenterol 21:9253–9261. https://doi.org/10.3748/wjg.v21.i31.9253
Aarnio M, Sankila R, Pukkala E et al (1999) Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer 81:214–218
Engel C, Loeffler M, Steinke V et al (2012) Risks of less common cancers in proven mutation carriers with lynch syndrome. J Clin Oncol 30:4409–4415. https://doi.org/10.1200/JCO.2012.43.2278
Dominguez-Valentin M, Sampson JR, Seppälä TT et al (2020) Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the Prospective Lynch Syndrome Database. Genet Med 22:15–25. https://doi.org/10.1038/s41436-019-0596-9
Lynch HT, Lanspa S, Shaw T et al (2017) Phenotypic and genotypic heterogeneity of Lynch syndrome: a complex diagnostic challenge. Fam Cancer. https://doi.org/10.1007/s10689-017-0053-3
Ligtenberg MJL, Kuiper RP, Chan TL et al (2008) Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet 41:112
Takahashi M, Furukawa Y, Shimodaira H et al (2012) Aberrant splicing caused by a MLH1 splice donor site mutation found in a young Japanese patient with Lynch syndrome. Fam Cancer 11:559–564. https://doi.org/10.1007/s10689-012-9547-1
Bianchi F, Raponi M, Piva F et al (2011) An intronic mutation in MLH1 associated with familial colon and breast cancer. Fam Cancer 10:27–35. https://doi.org/10.1007/s10689-010-9371-4
Cartegni L, Wang J, Zhu Z et al (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 31:3568–3571. https://doi.org/10.1093/nar/gkg616
Sharp A, Pichert G, Lucassen A, Eccles D (2004) RNA analysis reveals splicing mutations and loss of expression defects in MLH1 and BRCA1. Hum Mutat 24:272. https://doi.org/10.1002/humu.9267
Stella A, Shito K, Liu B et al (2001) A nonsense mutation in MLH1 causes exon skipping in three unrelated HNPCC families. Cancer Res 61:7020–7024
Tournier I, Vezain M, Martins A et al (2008) A large fraction of unclassified variants of the mismatch repair genes MLH1 and MSH2 is associated with splicing defects. Hum Mutat 29:1412–1424. https://doi.org/10.1002/humu.20796
Soukarieh O, Gaildrat P, Hamieh M et al (2016) Exonic splicing mutations are more prevalent than currently estimated and can be predicted by using in silico tools. PLoS Genet 12:1–26. https://doi.org/10.1371/journal.pgen.1005756
Thompson BA, Spurdle AB, Plazzer J-P et al (2013) Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet 46:107–115. https://doi.org/10.1038/ng.2854
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–423. https://doi.org/10.1038/gim.2015.30
Dominguez-Valentin M, Nakken S, Tubeuf H et al (2018) Identification of genetic variants for clinical management of familial colorectal tumors. BMC Med Genet 19:26. https://doi.org/10.1186/s12881-018-0533-9
Köger N, Paulsen L, López-Kostner F et al (2018) Evaluation of MLH1 variants of unclear significance. Genes Chromosom Cancer 57:350–358. https://doi.org/10.1002/gcc.22536
Dominguez-Valentin M, Evans DGR, Nakken S et al (2018) Genetic variants of prospectively demonstrated phenocopies in BRCA1/2 kindreds. Hered Cancer Clin Pract 16:4. https://doi.org/10.1186/s13053-018-0086-0
Vaccaro CA, López-Kostner F, Adriana DV et al (2018) From colorectal cancer pattern to the characterization of individuals at risk: Picture for genetic research in Latin America. Int J Cancer. https://doi.org/10.1002/ijc.31920
Dominguez-Valentin M, Nakken S, Tubeuf H et al (2018) Potentially pathogenic germline CHEK2 c.319+2T%3eA among multiple early-onset cancer families. Fam Cancer 17:141–153. https://doi.org/10.1007/s10689-017-0011-0
Baralle D, Lucassen A, Buratti E (2009) Missed threads. The impact of pre-mRNA splicing defects on clinical practice. EMBO Rep 10:810–816. https://doi.org/10.1038/embor.2009.170
Dominguez-Valentin M, Nilbert M, Wernhoff P et al (2013) Mutation spectrum in South American Lynch syndrome families. Hered Cancer Clin Pract 11:18. https://doi.org/10.1186/1897-4287-11-18
Rossi BM, Palmero EI, López-Kostner F et al (2017) A survey of the clinicopathological and molecular characteristics of patients with suspected Lynch syndrome in Latin America. BMC Cancer 17:623. https://doi.org/10.1186/s12885-017-3599-4
Carneiro da Silva F, de Ferreira JRO, Torrezan GT et al (2015) Clinical and molecular characterization of Brazilian patients suspected to have lynch syndrome. PLoS ONE 10:e0139753–e0139753. https://doi.org/10.1371/journal.pone.0139753
Della Valle A, Rossi BM, Palmero EI et al (2019) A snapshot of current genetic testing practice in Lynch syndrome: the results of a representative survey of 33 Latin American existing centres/registries. Eur J Cancer 119:112–121. https://doi.org/10.1016/j.ejca.2019.07.017
den Dunnen JT, Antonarakis SE (1999) Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 15:7–12
Valentin MD, Da Silva FC, Dos SEMM et al (2011) Characterization of germline mutations of MLH1 and MSH2 in unrelated south American suspected Lynch syndrome individuals. Fam Cancer 10:641–647. https://doi.org/10.1007/s10689-011-9461-y
Houdayer C, Caux-Moncoutier V, Krieger S et al (2012) Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum Mutat 33:1228–1238. https://doi.org/10.1002/humu.22101
Gaildrat P, Killian A, Martins A et al (2010) Use of splicing reporter minigene assay to evaluate the effect on splicing of unclassified genetic variants. In: Webb M (ed) Cancer susceptibility: methods and protocols. Humana Press, Totowa, pp 249–257
Ho SN, Hunt HD, Horton RM et al (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. https://doi.org/10.1016/0378-1119(89)90358-2
Pavicic W, Nieminen TT, Gylling A et al (2014) Promoter-specific alterations of APC are a rare cause for mutation-negative familial adenomatous polyposis. Genes Chromosom Cancer. https://doi.org/10.1002/gcc.22197
Casey G, Lindor NM, Papadopoulos N, Al E (2005) Conversion analysis for mutation detection in mlh1 and msh2 in patients with colorectal cancer. JAMA 293:799–809
Pagenstecher C, Wehner M, Friedl W et al (2006) Aberrant splicing in MLH1 and MSH2 due to exonic and intronic variants. Hum Genet 119:9–22. https://doi.org/10.1007/s00439-005-0107-8
Petersen SM, Dandanell M, Rasmussen LJ et al (2013) Functional examination of MLH1, MSH2, and MSH6 intronic mutations identified in Danish colorectal cancer patients. BMC Med Genet 14:103. https://doi.org/10.1186/1471-2350-14-103
Lefevre JH, Colas C, Coulet F et al (2010) MYH biallelic mutation can inactivate the two genetic pathways of colorectal cancer by APC or MLH1 transversions. Fam Cancer 9:589–594. https://doi.org/10.1007/s10689-010-9367-0
Maquat LE (1995) When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA 1:453–465
Silva FCC, Torrezan GT, Ferreira JRO et al (2017) Germline mutations in MLH1 leading to isolated loss of PMS2 expression in Lynch syndrome. Am J Surg Pathol 41:861–864. https://doi.org/10.1097/PAS.0000000000000827
Thompson BA, Martins A, Spurdle AB (2015) A review of mismatch repair gene transcripts: issues for interpretation of mRNA splicing assays. Clin Genet 87:100–108. https://doi.org/10.1111/cge.12450
Goldberg Y, Porat RM, Kedar I et al (2008) Mutation spectrum in HNPCC in the Israeli population. Fam Cancer 7:309–317. https://doi.org/10.1007/s10689-008-9191-y
Wolf B, Henglmueller S, Janschek E et al (2005) Spectrum of germ-line MLH1 and MSH2 mutations in Austrian patients with hereditary nonpolyposis colorectal cancer. Wien Klin Wochenschr 117:269–277
Burn J, Gerdes AM, MacRae F et al (2011) Long-term effect of aspirin on cancer risk in carriers of hereditary colorectal cancer: an analysis from the CAPP2 randomised controlled trial. Lancet 378:2081–2087. https://doi.org/10.1016/S0140-6736(11)61049-0
Sahashi K, Masuda A, Matsuura T et al (2007) In vitro and in silico analysis reveals an efficient algorithm to predict the splicing consequences of mutations at the 5′ splice sites. Nucleic Acids Res 35:5995–6003. https://doi.org/10.1093/nar/gkm647
van der Klift HM, Jansen AML, van der Steenstraten N et al (2015) Splicing analysis for exonic and intronic mismatch repair gene variants associated with Lynch syndrome confirms high concordance between minigene assays and patient RNA analyses. Mol Genet Genom Med 3:327–345. https://doi.org/10.1002/mgg3.145
Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3:285–298. https://doi.org/10.1038/nrg775
Krawczak M, Thomas NST, Hundrieser B et al (2007) Single base-pair substitutions in exon–intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum Mutat 28:150–158. https://doi.org/10.1002/humu.20400
Buratti E, Chivers M, Královičová J et al (2007) Aberrant 5′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res 35:4250–4263. https://doi.org/10.1093/nar/gkm402
Acknowledgements
We thank the patients and their families who participated in this work. We also want to thank Fabiana Alejandra Ferro, Pablo Germán Kalfayan and Juan Pablo Santino from Pro.Can.He for their support in Argentinean patient’s management and preparation of all the familial pedigrees, genetic variant analysis and IHC data analysis, respectively and the A.C. Camargo Biobank for sample processing. This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica [PICT-2017-3210 to C.V. and W.P.]; and the Instituto Nacional del Cáncer de Argentina [INC-Grant AFIV-2018/2020 to C.V. and W.P.]; and by Fundação de Amparo à Pesquisa do Estado de São Paulo [2014/50943-1 to DMC and GTT]. Moreover, this study was sponsored by the Groupement des Entreprise Françaises dans la Lutte contre le Cancer (Gefluc), and co-supported by the European Union and Région Normandie. Europe gets involved in Normandy with European Regional Development Fund (ERDF). This work was also supported by the Radium Hospital Foundation (Oslo, Norway), the Norwegian Cancer Society, contract 194751-2017 and Helse Sør-Øst (Norway).
Funding
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica [PICT-2017-3210 to C.V. and W.P.]; and the Instituto Nacional del Cáncer de Argentina [INC-Grant AFIV-2018/2020 to C.V. and W.P.]; and by Fundação de Amparo à Pesquisa do Estado de São Paulo [2014/50943-1 to DMC and GTT]. Moreover, this study was sponsored by the Groupement des Entreprise Françaises dans la Lutte contre le Cancer (Gefluc), and co-supported by the European Union and Région Normandie. Europe gets involved in Normandy with European Regional Development Fund (ERDF). This work was also supported by the Radium Hospital Foundation (Oslo, Norway), the Norwegian Cancer Society, contract 194751-2017 and Helse Sør-Øst (Norway).
Author information
Authors and Affiliations
Contributions
TAP, OS, MDV, PM, CAV, AM and WHP contributed to conception and design of the study, and wrote the manuscript. TAP, OS, MR, KA, FLK, GTT, DMC, ILON, TFB, TMBML, JCF, MBT, KAS, BMR, SAJ, JM, MDV, CAV, AM and WHP contributed to acquisition and analysis of data. TAP, OS, GTT, AM and WHP prepared the figures. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Consent to participate and for publication
Patients were informed about their inclusion into the registries and written informed consent was obtained from all participants during genetic counseling sessions.
Ethics approval
This study was approved by the institutional review board of the Hospital Italiano de Buenos Aires (Buenos Aires, Argentina), the A.C. Camargo Cancer Center (São Paulo, Brazil), the Federal University of Bahia (Bahia, Brazil) and the Clinica Las Condes (Santiago de Chile, Chile). All procedures followed were in accordance with the ethical standards of the Helsinki Declaration.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Piñero, T.A., Soukarieh, O., Rolain, M. et al. MLH1 intronic variants mapping to + 5 position of splice donor sites lead to deleterious effects on RNA splicing. Familial Cancer 19, 323–336 (2020). https://doi.org/10.1007/s10689-020-00182-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-020-00182-5