Abstract
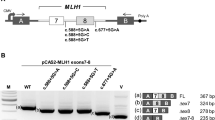
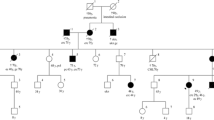
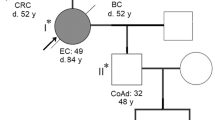
Germline mutations in the DNA mismatch repair (MMR) genes cause Lynch syndrome. Classification and interpretation of intronic variants, especially those outside the consensus ± 1 ~ 2 splice sites are challenging as it is uncertain whether such variants would affect splicing accuracy and efficiency. The assessment of the pathogenicity of splice site variants in MLH1 is further complicated by the various isoforms due to alternative splicing. In this report, we describe a 42-year-old female with Lynch syndrome who carries a germline variant, MLH1 c.678-3T>A, in the splice acceptor site of intron 8. Functional studies and semiquantitative analysis demonstrated that this variant causes a significant increase in the transcripts with exon 9 or exon 9 and 10 deletions, which presumably leads to premature protein truncation or abnormal protein. In addition, we also observed MSI-H and loss of MLH1 by IHC in patient’s tumor tissue. This variant also segregated with Lynch Syndrome related cancers in three affected family members. Based on these evidence, the MLH1 c.678-3T>A variant is considered pathogenic.




Similar content being viewed by others
References
Goecke T, Schulmann K, Engel C, Holinski-Feder E, Pagenstecher C, Schackert HK, Kloor M, Kunstmann E, Vogelsang H, Keller G, Dietmaier W, Mangold E, Friedrichs N, Propping P, Kruger S, Gebert J, Schmiegel W, Rueschoff J, Loeffler M, Moeslein G, German HC (2006) Genotype-phenotype comparison of German MLH1 and MSH2 mutation carriers clinically affected with Lynch syndrome: a report by the German HNPCC Consortium. J Clin Oncol 24(26):4285–4292. https://doi.org/10.1200/JCO.2005.03.7333
Vasen HF (2005) Clinical description of the Lynch syndrome [hereditary nonpolyposis colorectal cancer (HNPCC)]. Fam Cancer 4(3):219–225. https://doi.org/10.1007/s10689-004-3906-5
Cohen SA, Leininger A (2014) The genetic basis of Lynch syndrome and its implications for clinical practice and risk management. Appl Clin Genet 7:147–158. https://doi.org/10.2147/TACG.S51483
Das FC, Wernhoff P, Dominguez-Barrera C, Dominguez-Valentin M (2016) Update on hereditary colorectal cancer. Anticancer Res 36(9):4399–4405. https://doi.org/10.21873/anticanres.10983
Lynch HT, de la Chapelle A (2003) Hereditary colorectal cancer. N Engl J Med 348(10):919–932. https://doi.org/10.1056/NEJMra012242
Boland CR, Koi M, Chang DK, Carethers JM (2008) The biochemical basis of microsatellite instability and abnormal immunohistochemistry and clinical behavior in Lynch syndrome: from bench to bedside. Fam Cancer 7(1):41–52. https://doi.org/10.1007/s10689-007-9145-9
Peltomaki P, Vasen H (2004) Mutations associated with HNPCC predisposition—update of ICG-HNPCC/INSiGHT mutation database. Dis Mark 20(4–5):269–276
Thompson BA, Martins A, Spurdle AB (2015) A review of mismatch repair gene transcripts: issues for interpretation of mRNA splicing assays. Clin Genet 87(2):100–108. https://doi.org/10.1111/cge.12450
O’Leary E, Larson E, Lin AY, Reardon S, Lee P, Russell M, Melnitchouk N, Hart S, Ghassemi K, Hecht JR, Wainberg Z, Yoo J (2014) A multi-disciplinary cancer program enhances hereditary colorectal cancer detection. Am J Digest Dis 1(1):62–66
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Kohonen-Corish M, Ross VL, Doe WF, Kool DA, Edkins E, Faragher I, Wijnen J, Khan PM, Macrae F, St John DJ (1996) RNA-based mutation screening in hereditary nonpolyposis colorectal cancer. Am J Hum Genet 59(4):818–824
Genuardi M, Viel A, Bonora D, Capozzi E, Bellacosa A, Leonardi F, Valle R, Ventura A, Pedroni M, Boiocchi M, Neri G (1998) Characterization of MLH1 and MSH2 alternative splicing and its relevance to molecular testing of colorectal cancer susceptibility. Hum Genet 102(1):15–20
Charbonnier F, Martin C, Scotte M, Sibert L, Moreau V, Frebourg T (1995) Alternative splicing of MLH1 messenger RNA in human normal cells. Cancer Res 55(9):1839–1841
Trojan J, Zeuzem S, Randolph A, Hemmerle C, Brieger A, Raedle J, Plotz G, Jiricny J, Marra G (2002) Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology 122(1):211–219
Peasland A, Matheson E, Hall A, Irving J (2010) Alternative splicing of hMLH1 in childhood acute lymphoblastic leukaemia and characterisation of the variably expressed Delta9/10 isoform as a dominant negative species. Leuk Res 34(3):322–327. https://doi.org/10.1016/j.leukres.2009.08.015
Arnold S, Buchanan DD, Barker M, Jaskowski L, Walsh MD, Birney G, Woods MO, Hopper JL, Jenkins MA, Brown MA, Tavtigian SV, Goldgar DE, Young JP, Spurdle AB (2009) Classifying MLH1 and MSH2 variants using bioinformatic prediction, splicing assays, segregation, and tumor characteristics. Hum Mutat 30(5):757–770. https://doi.org/10.1002/humu.20936
Auclair J, Busine MP, Navarro C, Ruano E, Montmain G, Desseigne F, Saurin JC, Lasset C, Bonadona V, Giraud S, Puisieux A, Wang Q (2006) Systematic mRNA analysis for the effect of MLH1 and MSH2 missense and silent mutations on aberrant splicing. Hum Mutat 27(2):145–154. https://doi.org/10.1002/humu.20280
Casey G, Lindor NM, Papadopoulos N, Thibodeau SN, Moskow J, Steelman S, Buzin CH, Sommer SS, Collins CE, Butz M, Aronson M, Gallinger S, Barker MA, Young JP, Jass JR, Hopper JL, Diep A, Bapat B, Salem M, Seminara D, Haile R, Colon Cancer Family R (2005) Conversion analysis for mutation detection in MLH1 and MSH2 in patients with colorectal cancer. JAMA 293(7):799–809. https://doi.org/10.1001/jama.293.7.799
Bianchi F, Raponi M, Piva F, Viel A, Bearzi I, Galizia E, Bracci R, Belvederesi L, Loretelli C, Brugiati C, Corradini F, Baralle D, Cellerino R (2011) An intronic mutation in MLH1 associated with familial colon and breast cancer. Fam Cancer 10(1):27–35. https://doi.org/10.1007/s10689-010-9371-4
Takahashi M, Furukawa Y, Shimodaira H, Sakayori M, Moriya T, Moriya Y, Nakamura Y, Ishioka C (2012) Aberrant splicing caused by a MLH1 splice donor site mutation found in a young Japanese patient with Lynch syndrome. Fam Cancer 11(4):559–564. https://doi.org/10.1007/s10689-012-9547-1
Tournier I, Vezain M, Martins A, Charbonnier F, Baert-Desurmont S, Olschwang S, Wang Q, Buisine MP, Soret J, Tazi J, Frebourg T, Tosi M (2008) A large fraction of unclassified variants of the mismatch repair genes MLH1 and MSH2 is associated with splicing defects. Hum Mutat 29(12):1412–1424. https://doi.org/10.1002/humu.20796
Thompson BA, Spurdle AB, Plazzer JP, Greenblatt MS, Akagi K, Al-Mulla F, Bapat B, Bernstein I, Capella G, den Dunnen JT, du Sart D, Fabre A, Farrell MP, Farrington SM, Frayling IM, Frebourg T, Goldgar DE, Heinen CD, Holinski-Feder E, Kohonen-Corish M, Robinson KL, Leung SY, Martins A, Moller P, Morak M, Nystrom M, Peltomaki P, Pineda M, Qi M, Ramesar R, Rasmussen LJ, Royer-Pokora B, Scott RJ, Sijmons R, Tavtigian SV, Tops CM, Weber T, Wijnen J, Woods MO, Macrae F, Genuardi M (2014) Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet 46(2):107–115. https://doi.org/10.1038/ng.2854
Borras E, Pineda M, Brieger A, Hinrichsen I, Gomez C, Navarro M, Balmana J, Ramon y Cajal T, Torres A, Brunet J, Blanco I, Plotz G, Lazaro C, Capella G (2012) Comprehensive functional assessment of MLH1 variants of unknown significance. Hum Mutat 33(11):1576–1588. https://doi.org/10.1002/humu.22142
Tricarico R, Kasela M, Mareni C, Thompson BA, Drouet A, Staderini L, Gorelli G, Crucianelli F, Ingrosso V, Kantelinen J, Papi L, De Angioletti M, Berardi M, Gaildrat P, Soukarieh O, Turchetti D, Martins A, Spurdle AB, Nystrom M, Genuardi M, InSi GHTVIC (2017) Assessment of the InSiGHT interpretation criteria for the clinical classification of 24 MLH1 and MSH2 gene variants. Hum Mutat 38(1):64–77. https://doi.org/10.1002/humu.23117
Funding
This study was funded by Department of Pathology, Memorial Sloan Kettering Cancer Center.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Dr. Cadoo reports institutional support for therapeutic clinical trial, travel and expenses from Astra Zeneca, institutional support for therapeutic clinical trial from Syndax Pharmaceuticals, personal fees and other from Tessaro, personal fees from OncLive, outside the submitted work. Dr. Zhang: honoraria (future technology research LLC, BGI, Illumina); honoraria and travel and accommodation expenses (Roche Diagnostics Asia Pacific). Family member has a leadership position and ownership interest of Shanghai Genome Center. The other authors do not have any conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yang, C., Sheehan, M., Borras, E. et al. Characterization of a germline splice site variant MLH1 c.678-3T>A in a Lynch syndrome family. Familial Cancer 19, 315–322 (2020). https://doi.org/10.1007/s10689-020-00180-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-020-00180-7