Abstract
This study aimed to identify wild barley alleles controlling grain size and weight with the potential to improve barley yield in Australia and worldwide. The HEB-25 nested association mapping population was used, which samples 25 different wild barley accessions in a ‘Barke’ genetic background. The HEB-25 population was evaluated in field conditions at Strathalbyn in South Australia in 2015 and 2016. Seven yield component traits reflecting ear length, grain number per ear and grain dimension were measured. Among 114 quantitative trait loci (QTL) identified for the seven traits in both years, many co-localise with known genes controlling flowering and spike morphology. There were 18 QTL hotspots associated with four loci or more, of which one at the beginning of chromosome 5H had wild alleles that increased both grain number per ear and thousand-grain weight. A wide range of effects was found for wild alleles for each trait across all QTL identified, providing a rich source of genetic diversity that barley breeders can exploit to enhance barley yield.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Barley is one of the most important cereal crops globally and is ranked fourth in the world in terms of quantity produced and area of cultivation. (Capettini et al. 2010; Zhou 2010). It is also the second most important crop in Australia, with average grain production of about 9 million tonnes per annum, 70% of which is exported (ABARES 2019). Australia makes up over 30% of the world malting barley trade and approximately 20% of the world feed barley trade (Barley 2022). Although barley yield in Australia increased from 1.2 t/ha in 1969 to 2.5 t/ha in 2015, this rate of genetic gain is still slower than those of other countries especially the European Union (EU) (2 to 5–8 ton/ha) (Schils et al. 2018; Spragg 2016). This is probably caused by low soil fertility, lower and highly variable precipitation with many stresses that reduce productivity in Australia (Hochman et al. 2013; Richards 1991; Van Gool and Vernon 2006). Thus, breeding for higher yield in Australia is a challenging task as grain yield has a relatively low heritability due to the high GEI (genotype × environment interaction) (Slafer 2003). Nevertheless, there have been continuous efforts spent to increase barley grain yield in this highly variable environment (Brown et al. 1988; Donald 1979; Eagles and Moody 2004; Finlay and Wilkinson 1963).
The most significant breakthrough in barley yield improvement was achieved by the introduction of Green Evolution genes sdw1.d, sdw1.c (originally named denso), uzu1.a, and erectodies (ari-e.GP), which resulted in shorter plants, better lodging resistance, and higher harvest index, thus leading to higher yield (Jia et al. 2016; Mickelson and Rasmusson 1994; Nadolska-Orczyk et al. 2017). However there is evidence that the best alleles conditioning the short stature in barley tend to become fixed in modern barley breeding germplasm (Jia et al. 2009). In addition, some of these genes (i.e. denso and uzu1.a) also were reported to associate with unwanted traits such as lowered malt quality or temperature sensitivity (Dockter et al. 2014; Hellewell et al. 2000; Wang et al. 2010b), and therefore, there were attempts to replace them with other plant height-conditioning genes (Dockter et al. 2014).
The other promising avenue employed to improve yield is to improve yield component traits such as number of spike/plant (or tiller number), grain number per ear, and thousand-grain weight (Jedel and Helm 1994; Peltonen-Sainio et al. 2007). Depending on the genotypes and the environments tested, the interaction among these three components was found to be either negative (Bulman et al. 1993; Hadjichristodoulou 1990), positive (Saade et al. 2016), or both, but depending on the environment (Markova et al. 2015; Wiegmann et al. 2019). This indicates that yield components also have strong GEI, and there is plasticity in the yield components to compensate for yield reduction (Sadras and Rebetzke 2013). Nevertheless, improving one trait without compromising one another was feasible (Griffiths et al. 2015; Zhou et al. 2016). Genotypic selection for yield component quantitative trait loci (QTL) in tandem with phenotypic yield selection may be more beneficial to improving yield than yield testing alone. QTL affecting the three aforementioned yield components have been defined and mapped in barley (Cu et al. 2016; Maurer et al. 2016; Mikołajczak et al. 2016; Sharma et al. 2018; Walker et al. 2013; Wang et al. 2019; Xu et al. 2018; Zhou et al. 2016). In recent studies, many “hotspots”, QTL regions controlling multiple traits, were identified. In a few cases, these hotspots reside near key genes controlling plant stature, flowering time, and spike morphology, such as vrs1, nud, VRN-H3 and sdw1/denso (Wang et al. 2019). In other cases, they co-locate with barley orthologs of genes controlling grain size and weight in other cereal species such as rice, maize, and wheat, demonstrating the conservation of gene function within the cereal family. Functional genomics and analysis of natural or induced mutants have revealed genes involved in spike morphology, such as Vrs1 (Komatsuda et al. 2007), Vrs5 (Ramsay et al. 2011), INT-C/HvTB1 (Lundqvist 1997), HvAP2 (Houston et al. 2013), and HvCKX (Zalewski et al. 2010, 2014, 2012) determine yield components and grain yield.
For a specific trait, the genetic improvement of crop plants is accomplished by stacking and accumulating favourable alleles within gene pools. In barley, domesticated barley (H. vulgare L. ssp. vulgare) and wild barley (H. vulgare L. ssp. spontaneum) constitute two primary sources of advantageous alleles in breeding (Wendler et al. 2014). Beneficial alleles from wild barley have been repeatedly used to improve yield in elite cultivars (Kalladan et al. 2013; Lakew et al. 2013; Nevo and Shewry 1992). To capture a wide range of genetic diversity from wild barley for better identification of beneficial alleles, a Nested Association Mapping (NAM) population was created from crossing 25 genetically diverse wild barley accessions originating from the Fertile Crescent region to the malting cultivar ‘Barke’ (Maurer et al. 2015). NAM populations combine the advantages and minimise the disadvantages of two traditional methods for identifying quantitative trait loci, including bi-parental and association mapping populations. NAM populations have higher allelic diversity and higher recombination events than biparental populations, decreasing the population structure's confounding effect seen in the association mapping populations (Gage et al. 2020; Hu et al. 2018; Kitony et al. 2021). HEB-25 population (HEB stands for Halle Exotic Barley) has been utilized to study flowering time, salinity and drought tolerance, grain size and weight and disease resistance in barley in a wide range of environments across the globe (Büttner et al. 2020; Herzig et al. 2018; Maurer et al. 2015; Maurer et al. 2016; Pham et al. 2019; Saade et al. 2016; Sharma et al. 2018; Vatter et al. 2017, 2018). This population harbours a reservoir of beneficial alleles that can be exploited for variety development and yield improvement in previously tested environments. In 2020, the HEB-25 population was evaluated and screened to identify QTL relating to phenology in Charlick, South Australia (Pham et al. 2020). Here, we report on the results of a Genome-Wide Association Study (GWAS) conducted for spike morphology-related traits from the same field trials conducted by Pham et al. (2020) that might contribute alleles to increase yield in environments such as those in southern Australia.
Materials and methods
The plant materials
The wild barley HEB-25 NAM population was developed by crossing and then backcrossing 25 diverse wild barley accessions (Hordeum vulgare ssp. spontaneum and agriocrithon) to the cultivar Barke. Details about the population design and development were described previously (Maurer et al. 2015). In summary, there were 25 subfamilies, with the number of lines within each family in the range from 22 (family no.18) to 75 (family no.3) (Maurer et al. 2015, Additional file 1). On average, there were 52 lines per family, with each BC1S3 plant hadving an expected segregation of 71.875% homozygous Barke loci, 21.875% homozygous wild barley loci, and 6.25% heterozygous loci. The lines sown in 2015 and 2016 were BC1S3:8 and BC1S3:9 lines, respectively.
Genotyping of NAM lines
The population was genotyped with the Illumina iSelect 50 K chip (Bayer et al. 2017), and a total of 32,995 SNPs meeting the quality criteria (polymorphic in at least one HEB family, < 10% failure rate, < 12.5% heterozygous calls) were utilised for GWAS in this study.
A quantitative identity by state (IBS) approach described by Maurer et al. (2015) was used to define the SNP matrix. Missing SNPs were imputed using the mean score of polymorphic flanking markers (matrix E, Maurer & Pillen, 2019, https://doi.org/https://doi.org/10.5447/ipk/2019/20).
HEB-25 field trials
Field trials were conducted at Charlick, Strathalbyn in South Australia in 2015 (− 35° 19′ 20″ N, 138° 53′ 24″ E) and 2016 (− 35° 19′ 22″ N, 138° 52′ 56″ E) with 941 and 1294 lines of the HEB-25 population respectively. The lines tested in 2015 were a subset of the population containing 18 families, of which seeds were available for testing at the time of sowing in 2015. The experiment design was fully described by Pham et al. (2020). In summary, the experiment design for 2015 was an unreplicated augmented block design with lines grouped by family. There were 1176 plots arranged into 12 bays × 98 columns with two plots for check varieties per column. For 2016, a partially replicated randomized design was implemented with 147 out of 1294 lines replicated, and the same check lines sown in 2015 were used again at a frequency of one check plot every 10 test lines. Check lines used in both years included ten Australian varieties (Admiral, Capstan, Commander, Compass, Fleet, Flagship, Hindmarsh, Gairdner, Navigator, and Keel), and the parental line Barke. Plots in 2015 and 2016 were 2.24 m2 doubled-rows with 3.20 m in length and separated by 0.5 m to reduce competition between plots. Two rows within a plot were 0.20 m apart.
The sowing and harvesting dates for the 2015 trial were June 16th and December 15th, respectively. The sowing and harvesting dates for the 2016 trial were May 18th 2016, and December 20th 2016, respectively.
Phenotypic data
Seven traits measured in this study included ear length (EL), awn length (AL), grain number per ear (GPE), grain area (GA), grain width (GW), grain length (GL), thousand-grain weight (TGW). For the first three traits, phenotypic values for each genotype were calculated as an average of measurements taken from 15 randomly chosen ears per plot. Ear length was measured in cm from the bottom to the top of the ear, and awn length was measured in cm from the top of the ear to the tip of the awn. Grain number per ear (GPE) was measured as total grain counts from each ear. The latter four were measured using the 150–300 seeds using GrainScan software (Whan et al. 2014) and expressed in mm for GL and GW, mm2 for GA and gram for TGW.
Within and between year phenotypic analysis
Within each of the years, the analysis of individual grain traits was conducted using a linear mixed model (LMM) that appropriately partitioned and accounted for all genetic and non-genetic sources of variation (Pham et al. 2020) specified as the following:
If \({\varvec{y}}=\left({y}_{1}, \dots , {y}_{n}\right)\) are a vector of \(n\) phenotypic trait responses, then the LMM was defined as
where \({\varvec{X}}{\varvec{\beta}}\) was the fixed component of the model and contained a population type factor to estimate the overall mean of the HEB-25 population as well as separate means for each of the parents and controls involved in the experiment. This component also contained terms to model distinct spatial trends such as linear column or row effects if present in the field. The term \({\varvec{Z}}{\varvec{u}}\) was the random component containing factors to model possible sources of non-genetic variation including non-linear row or column effects. This component may have also contained terms to model potential outliers using the methodology outlined by Gumedze et al. (2010). Additional extraneous spatial variation was captured with the residual model error term, \({\varvec{e}}\), and was assumed to be distributed \({\varvec{e}} \sim N(0, {\varvec{R}}\)) where \({\varvec{R}} = \sigma^{2} {{\varvec{\Sigma}}}_{r} \otimes {{\varvec{\Sigma}}}_{c}\) is a separable correlation structure with \({{\varvec{\Sigma}}}_{r}, {{\varvec{\Sigma}}}_{c}\) parameterized as an auto-regressive structure of order 1 in the row and column direction respectively. The underlying genetic variation of the HEB-25 lines was modelled using the random component term \({{\varvec{Z}}}_{g}{\varvec{g}}\) where the genetic random effects, \({\varvec{g}}\), are an \(r\) length vector and assumed to be distributed \({\varvec{g}} \sim N(0, {\sigma }_{g}^{2}{{\varvec{I}}}_{r})\). This assumes a common genetic variance across the HEB-25 population. Under this LMM structure the effects, \(\left({\varvec{u}}, {\varvec{g}}, {\varvec{e}}\right)\), were considered to be mutually independent. From each of the fitted models, the vector of best linear unbiased predictions (BLUPs) \(\widetilde{{\varvec{g}}}=({\widetilde{g}}_{1}, \dots , {\widetilde{g}}_{r})\) of the HEB-25 lines, as well as their prediction error variances, were used to calculate broad sense generalized heritabilities with the formula developed by Cullis et al. (2006), namely
where \(PE{V}_{ave}\) is the average of the prediction error variances of all elementary contrasts between the progeny lines and \({\widehat{\sigma }}_{g}^{2}\) is a REML estimate of the genetic variance of the progeny obtained from the fitted model.
To understand the genetic relatedness of the traits between the 2 years, (1) was extended to a multi-year LMM. For each trait, the fixed component of the multi-year LMM consisted of an interaction of a two-level year factor with a population factor to ensure HEB-25 progeny means, parents and controls were estimated independently for each year. Extraneous sources of environmental variation modelled with fixed and random terms in the single year models, were also appropriately added to the multi-year LMM. The multi-year LMM residual was assumed to be distributed \({\varvec{e}} \sim N\left( {0, \oplus_{i = 1}^{2} {\varvec{R}}_{i} } \right)\) where \(\oplus_{i = 1}^{2} {\varvec{R}}_{i} = {\text{diag}}\left( {{\varvec{R}}_{i} } \right)\) (Butler et al. 2018) with \({\varvec{R}}\) defined previously. Most importantly, the random genetic effects are assumed to have a multiplicative structure with distribution \({\varvec{g}} \sim N\left( {0,{\varvec{G}} \otimes {\varvec{I}}_{r} } \right)\) where \({\varvec{G}}\) is a 2 × 2 correlation matrix with diagonal elements reflecting the underlying genetic variation of the HEB-25 lines in each year and off diagonal covariance reflecting the genetic relationship of the lines between years.
Computations and heritability
All phenotypic models were fitted using the LMM software ASReml-R (Butler et al. 2018), available as a package in the R statistical computing environment. ASReml-R uses a residual maximum likelihood approach toestimate mean and variance parameters (Patterson and Thompson 1971).
For individual traits analysed using the LMM, broad sense generalised heritabilities were calculated using the formula Cullis et al. (2006) described.
Genome-wide association study
QTL detection and cross-validation were conducted as described by Büttner et al. (2020). The detection rate was calculated to represent validity and significance, and each marker detected in at least 20 cross-validation runs was declared significantly associated with the trait. Significant marker-trait associations were grouped to a single QTL if the significant SNPs were linked by less than 5 cM and expressed the same direction of additive effects, i.e. both exotic alleles increased or decreased the trait of interest. We applied the cumulation method to estimate a parent-specific QTL effect, as presented in Maurer et al. (2017). This procedure was conducted within each cross-validation run, and their mean of them was taken as the final parent-specific QTL effect estimate. The required genetic positions of 50 k markers were estimated as described in Büttner et al. (2020). All SNPs from the respective QTL interval were fitted in a linear model to estimate the QTL’s explained phenotypic variance (Vp) in the whole dataset.
Specifically, preceding marker-based analysis, the BLUPs for individual traits within each year were de-regressed using the formula described by Garrick et al. (2009), namely
where \({\widetilde{{\varvec{g}}}}_{{\varvec{i}}}\) and \({\varvec{P}}{\varvec{E}}{{\varvec{V}}}_{{\varvec{i}}}\) is the BLUP and prediction error variance of the \({\varvec{i}}\)th line respectively.
For each set of de-regressed BLUPs, a GWAS was conducted using a two stage multiple regression procedure similar to the methods described by Liu et al. (2011) and Maurer et al. (2016). In the first stage a set of SNP co-factors are sought of from an initial saturated additive SNP model specified as
where \({{\varvec{\tau}}}_{{\varvec{p}}}\) was a vector of fixed effect parameters used to estimate the NAM sub-population means and \({{\varvec{X}}}_{{\varvec{p}}}\) was the associated indicator matrix that maps trait responses to the appropriate sub-population. In this saturated SNP model \({{\varvec{M}}}_{{\varvec{i}}}\) was the \({\varvec{i}}\)th genetic marker covariate containing quantitative allelic values spanning the NAM genotypes and \({{\varvec{q}}}_{{\varvec{i}}}\) is its associated effect size. The model error, \({{\varvec{e}}}^{\boldsymbol{*}}\) represents residual genetic error and is assumed to be distributed \({{\varvec{e}}}^{\boldsymbol{*}}\boldsymbol{ }\sim \boldsymbol{ }{\varvec{N}}(0,\boldsymbol{ }{{\varvec{\sigma}}}^{2}{{\varvec{I}}}_{{\varvec{r}}})\). To determine the set of SNP co-factors, a forward–backward stepwise regression approach was implemented where inclusion or exclusion of individual SNPs was determined through minimising the Bayesian Information Criterion. From the determined set of \({\varvec{c}}\) SNP co-factors, a genome wide scan is then conducted with each marker individually added to a multiple regression model of the form
where \({{\varvec{M}}}_{{\varvec{k}}}\) is the marker being assessed and co-factors that were less than 1 cM from the marker being assessed were excluded from the co-factor set. The Bonferroni-Holm method (Holm 1979) was used to provide a family-wise adjusted \({\varvec{p}}\)-value for multiple testing of marker-trait associations with significant markers accepted if \({{\varvec{p}}}_{{\varvec{B}}{\varvec{O}}{\varvec{N}}-{\varvec{H}}{\varvec{O}}{\varvec{L}}{\varvec{M}}}<0.05\). The approximate proportion of genetic variance explained by a marker was determined by estimating \({{\varvec{R}}}^{2}\) after modelling the marker solely in a linear model.
Candidate genes for QTL detected by GWAS were identified using the BARLEYMAP pipeline, MorexV3 map (Cantalapiedra et al. 2015). In addition we compared the genomic position of the detected QTL with the position of 164 barley orthologs with the cereal genes for grain size and weight reported by Wang et al. 2019. Genes were suggested as candidates if they were within 4 cM upstream or downstream of a QTL, reflecting the LD decay of 7.85 cM reported in HEB-25 by Vatter et al. (2017).
Results
Trait variation, trait heritability, and multiyear relationship
Among the investigated traits, large ranges in phenotypic values were observed, with the maximum values often 2-3X larger than the minimum values. (Fig. 1 and Table 1). HEB-25 lines surpassing the recurrent parent ‘Barke’, and ten check varieties were observed for all measured traits except for TGW, GW, and GA in 2015 and AL, GA, and GW in 2016 (Fig. 1 and Online Resource 1).
When a Student t-test was used, a significant difference between the 2 years was observed for all traits measured (P < 0.05, data not shown). The mean trait values for AL, EL, GA, and GL in 2015 were higher than those in 2016, while the reverse trend was found for GPE, TGW and GW. The significant difference between the 2 years for the seven traits measured was probably attributable to the difference in cultural practice, environment and genetic composition between 2 years. The trial in 2015 was sown 1 month later than the usual sowing date for the location (mid-May) as there was little rain to support seed growth in May 2015. Growing season rainfall (GSR) of 2015 and 2016 at the tested location was 229 mm and 502 mm, respectively, representing a reduction of 26% and an increase of 60% compared to the 26-year average for the location (Online Resource 2). Finally, seeds of only 18 families were sown in 2015 as the other seven families were still under assessment in quarantine.
In general, heritabilities were relatively high, ranging from 0.49 to 0.84 (Table 2). Heritabilities were higher for 2016 compared to 2015, possibly due to the smaller number of lines tested in 2015 compared to 2016. The lowest heritability was found for GPE in 2015 (0.49), and the highest heritability was found for GW in 2016 (0.87). The difference in magnitude of heritability between the 2 years was greatest for GPE and GW.
Although the heritabilities of the traits were consistent between 2 years, the within-year genetic correlations ranged from 0.57 to 0.69 except for GW (0.84), reflecting a reduced genetic similarity in each of the studied traits between the years (Table 2). Taking everything into consideration, traits within each year were individually processed for further analysis.
Within-year trait relationships
Correlations among seven measured traits are shown in Fig. 2. In both years, TGW had a strong positive correlation with GW and GA and weaker positive correlations with GL and AL. There was a correlation with GPE and EL in 2016 only.
Correlation matrices for seven traits measured from field trials from 2015 to 2016. Correlation matrices for seven traits in 2015 and 2016. In the following plots, the distribution of each variable is shown on the diagonal. The bivariate scatter plots with a fitted line are displayed on the bottom of the diagonal. The value of the correlation plus the significance level as stars are displayed on the top of the diagonal. Each significance level is associated to a symbol: p-values (0, 0.001, 0.05, 0.01) < = > symbols (“***”, “**”, “*”)
GA correlated positively and strongly with GL, GW, and TGW. Both AL and GL showed a negative correlation with GPE. AL had a low but significant positive correlation with TGW, GA and GL but not with GW. Although slightly differing in magnitude, a similar correlation trend among these variables was also observed in the field trials in Germany and Scotland for the HEB-25 population, as shown by Sharma et al. (2018).
GWAS results
The summary and full GWAS output of seven traits in both years were listed in the Table 2 and Online Resource 3, respectively.
GWAS-AL: there were 13 and 14 QTLs identified for awn length in 2015 and 2016, respectively, with four common QTLs between 2 years. The QTLs explained that the most phenotypic variation (Vp) in 2015 and 2016 was QAl.HEB-25-3H.2 (43.5 cM, 15%) and QAl.HEB-25-1H.3 (97.9 cM, 20%), respectively.
GWAS-EL: there were 10 and 13 QTLs identified for EL in 2015 and 2016, respectively, with three common QTLs between 2 years. The QTLs that explained the most Vp in 2015 and 2016 were QEl.HEB-25-2H.1 (63.55 cM, 11.4%) and QEl.HEB-25-3H.1 (40.7 cM, 21.3%), respectively. Among the three common QTLs for EL, the QTLs QEl.HEB-25-4H.1 had wild alleles that increased EL in both years, while the remaining two QTLs on chromosomes 5H and 7H had wild alleles with a mixed effect in both years.
GWAS-GPE: there were 7 and 14 QTLs detected for grain number per ear for 2015 and 2016, respectively, with two QTLs shared between 2 years. Both common QTLs had wild alleles from all families that reduced GPE. The QTLs explained that the largest Vp were QGpe.HEB-25-7H.1 (11.3%) and QGpe.HEB-25-2H.1 (21.7%) in 2015 and 2016, respectively. The QTLs at which wild alleles increased GPE the most were QGne.HEB-25-3H.3 (52.6 cM) in 2015 (up to 1.4 grain on average) and QGne.HEB-25-6H.1 (116.75 cM) on 2016 (increased GPE up to 2.3 grain in average) (Fig. 3).
GWAS-TGW: 8 and 23 QTLs were identified for TGW in 2015 and 2016, respectively TGW QTLs were mapped to all chromosomes. The QTLs explained that the largest Vp were QGpe.HEB-25-4H.4 (20.6%) and QGpe.HEB-25-6H.1 (20.9%) in 2015 and 2016, respectively. There were three common QTLs between the 2 years, with the two QTLs QTgw.HEB25-6H.1 and QTgw.HEB25-7H.2 having wild allele reduced TGW in both years in most of the HEB-25 families. In contrast, the common QTLs QTgw.HEB25-5H.1 increased TGW in both years in all families. The QTLs that increased TGW the most was QTgw.HEB25-6H.3 at which wild alleles from only family no. 25 increased TGW to 5.35 g. However, it was only detected in 2016, and alleles from all other families except no.25 reduced TGW.
GWAS-GL, GW, GA: there were 13 QTLs detected for GL in either 2015 or 2016, with only one common QTLs between 2 years. These QTLSs were mapped to all chromosomes and individually explained 1–35.2% of Vp. There were 12 and 16 QTLs detected for GA in 2015 and 2016, respectively, with five shared between 2 years. An individual QTLs explained from 5.9 to 35.5% of the phenotypic variance. The QTLs explained that the most Vp in both years for GL and GA was QGl.HEB25-1H.1, which co-localized with the thresh-1 locus (Schmalenbach et al. 2011).
There are 17 and 21 QTLs detected for GW in 2015 and 2016, respectively, with six shared between 2 years. These QTLSs were mapped to all chromosomes, and an individually explained 6.2 to 27.1% of the phenotypic variance The QTLs explained that the most Vp for GL and GA in both years was QGl.HEB25-1H.1, which co-localized with the HvAPO2/BFL locus.
Hotspot for grain size and weight
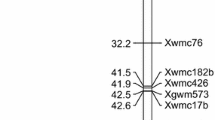
When all trait QTLs detected were plotted on the barley genetic map, several regional ‘hotspots’ were associated with multiple traits. There were 18 ‘hotspots’, defined as a window within 7.85 cM where four or more QTLs coincide, as this window was reported to be the linkage disequilibrium decay for this population by Vatter et al. (2017) (Fig. 4). There were three hotspots in each of the five chromosomes 1H, 2H, 4H, 5H and 7H, two hotspots in chromosome 3H and one in chromosome 6H. The hotspot 1_2 was found to associate with five traits, three of which had QTLs within this hotspot in both years. Another hotspot, 3_1, was linked with QTLs of six traits measured in this study. The rest of the hotspots associated with three to four traits.
Barley grain and ear dimension QTLs–genetic distributions and overlaps. The genetic positions of all QTLs identified here were based on the genetic map developed by Maurer et al. (2015) and (Büttner et al. 2020). The QTLs affecting four or more traits were grouped into QTL hotspots which shown in the rectangular boxes. Also plotted are genetic map locations for barley orthologs of known cereal grain trait genes (Wang et al. 2019) and other developmental genes known to affect yield in barley. Bars indicate the detection rate of each QTL detected for each trait, green and orange represents 2015 and 2016, respectively
Discussion
Phenotypic variation compared to the control varieties reveals promising candidate lines for breeding
For all seven traits except for GW, there were individuals with phenotypic values greater than most of the ten check varieties (Online Resource 1), implying the potential for use of the HEB-25 wild barley alleles in breeding to improve yield and yield component traits in Australian environments. Two HEB-25 lines, HEB-16-121 and HEB-16-144, had significantly higher GPE (up to 10 grains/ear) while having TGW equivalent to Compass, the current benchmark variety for high TGW and plumpness in Australia. In addition, line HEB-22-118 had TGW and GPE greater than Compass in both years, and its tiller number was equal to that of Compass in the wetter year 2016 (Online Resource 4, Pham et al. 2020). The correlation between GPE and TGW was found to be very low (< 0.1) for both years in the field trials, which was similar to findings from the salt stress study by Saade et al. (2016) and Sharma et al. (2018). The independent relationship of these traits in the HEB-25 population would be advantageous as beneficial alleles conditioning high GPE and high TGW can be combined into one genetic background without worrying about the trade-off effect between these two traits.Detection of a QTL is merely the result of statistical work using the association between genotype and phenotype. Therefore, the positive effect of identified beneficial alleles needs to be further validated in different genetic backgrounds using either biparental populations or a panel of diverse germplasm lines (Langridge et al. 2001; Pu-yang et al. 2022; Zhang et al. 2019). The beneficial wild allele conditioning increased GPE and TGW in the above lines. It was introgressed into three Australian cultivars, Compass, LaTrobe and Granger, via backcrossing and Kompetitive allele specific PCR (KASP) markers. We hypothesize that some progenies with transgressive segregants from these crosses will surpass the yield of the recurrent parental elite cultivars Compass, LaTrobe, and Granger. Both subsequent genomic and phenotypic selection will enable the identification of lines with potential high yielding from these crosses. Ultimately, field trials need to be conducted in the future to validate the effectiveness and drawbacks (i.e. linkage drag) of these beneficial wild alleles to yields and yield components.
Hotspots with beneficial wild alleles detected in this study and possible candidate genes
Eight hotspots were found to be common when the locations of our 18 hotspots were aligned to those of 14 hotspots detected for grain size and weight when the HEB-25 was evaluated in Germany and Scotland (Sharma et al. 2018). Among these typical hotspots, few co-localize with genes regulating flowering time/development, including Ppd_H2, HvELF3, HvCEN, HvCO1, and the remaining located closely to barley orthologs of cereal genes, including SRS3, GSK2, qGL3/GL3.1, and GW2 (Xu et al. 2018).
In this study, only three of the 16 hotspots (2_1, 4_1, and 5_1) had wild alleles, which positively affected on the traits measured, while others had a mixed effect or only negative effect. Wild alleles at these three hotspots increased GPE up to 0.9 grains/ear (hotspot 4_1 in family no.1, equivalent to 4.1% increase) and TGW up to 2 g (hotspot 2_1 in family no. 4, equivalent to 5.5% increase) compared to the Barke alleles. At the hotspot 5_1, four important yield component traits (GPE, TGW, GA and GW) were improved by the wild alleles from all families. It is noteworthy that the beneficial QTLs detected for GPE at these hotspots 4_1 and 5_1 were not found in the European counterpart study, signifying the value of this study that not only we can learn about the essential core loci/genes underlying one trait across environments but also those that are useful yet very environment-specific. Another advantage of these hotspots is that they reside near the end of the chromosome with a higher recombination rate, thus greatly facilitating recombination-based breeding.
Among these three hotspots, only one (4_1) has a potential candidate gene, LONELY GUY (LOG), which is a gene that has a direct role in the activation of cytokinins (Kuroha et al. 2009; Tokunaga et al. 2012). Rice plants with mutant log had abnormal branching, reduced number of floral organs and inflorescence complexity (Kurakawa et al. 2007).
When markers linked with the QTLs within the hotspot 2_1 and 5_1 were used to search for candidate genes within 4 cM upstream and downstream of these markers, 28 genes and 39 genes with high confidence were detected, respectively (Online Resource 5). For the hotspot 2_1, it is difficult to pinpoint a potential candidate gene for this region. In contrast, four candidate genes such as sucrose-phosphatase 1 (HORVU5Hr1G000580), ethylene receptor 1 (HORVU5Hr1G000590), MAD-box transcription factor (HORVU5Hr1G000370 or HORVU5Hr1G000480) could be the most promising candidate genes for the hotspot 5_1 as these were shown to hold a critical role to the grain filling process and grain weight (Jiang et al. 2011; Jiang et al. 2015; Luo et al. 2019; Paul et al. 2020; Radchuk et al. 2021; Wuriyanghan et al. 2009; Chen et al. 2016; Yang et al. 2012).
Comparison to QTLs detected in other studies for GPE and TGW
For GPE: across the four global field trials with the HEB-25 population conducted in Germany, Scotland, United Arab Emirates, and Australia, QTLs located near five loci including HvCEN (2H-56 cM), Vrs1 (2H-80 cM), Vrs4/btr1/btr2 (3H-46 cM), LOG (4H-1 cM), and VRN-H3 (7H-34 cM) were always detected regardless of the environment (Sharma et al. 2018; Saade et al. 2016). When GPE was mapped using materials with less complex genetic structure (association panel, double haploid populations, or introgression lines from wild barley), fewer QTLs linking with previously known genes controlling grain number observed, such as loci near Vrs1 and VRN-H3 were reported by Li et al. (2006); Ren et al. (2013), or only Vrs1 (Wang et al. 2016), HvCEN and Vrs1 (Honsdorf et al. 2017, Xu et al. 2018), or Ppd-H1, VRN2 and VRN-H3 (Wang et al. 2010a) were detected for GPE. Vrs4 was reported to regulate Vrs1 to control spikelet determinacy and morphology and indirectly control LOG-like gene expression (Koppolu et al. 2013). Furthermore, HvCEN was reported to interact with HvFT1 (VRN-H3) to regulate floral development and, thus, indirectly control GPE (Bi et al. 2019; Loscos et al. 2014). The consistent result in finding the five loci associated with GPE in experiments across the globe demonstrates the power of the HEB-25 population to detect essential genes/loci regulating GPE due to its intrinsic wide genetic variation.
In this study, the two QTLs at which wild alleles increased GPE the most were QGpe.HEB25-4H.3 (4H-43.5 cM, family no.16) and QGpe.HEB25-6H.1 (6H-117 cM, family no. 17 and 25). The former QTL was not detected when HEB-25 was evaluated in the European environments where the latter was only detected in Scotland, with much lower effect. Both of these two QTLs were detected in Dubai (UAE) by Saade et al. (2016) for GPE, but in that study, the former did have the largest effect while the latter QTL expressed a negative effect. In contrast, the QTL with the biggest GPE promoting effect in the counterpart European study on chromosome 5H at 165 cM (family no.18) was not detected for GPE in South Australian environment. Thus, this study provides another piece of the puzzle to help barley breeders better understand the location-specific effect of the wild barley alleles governing GPE in the HEB-25 population. In this study, the markers used to map yield component traits in the HEB-25 population were 6X more than those used in previous GWAS studies by Saade et al. (2016) and Sharma et al. (2018). The markers identified in this study, thus, are expected to be closer to the actual gene(s) than those identified in the previous study. For example, the marker associated with GPE and located near Vrs1 genes in this study, JHI-Hv50k-2016-107351, was 0.35 cM (777 kb in physical distance) from the actual Vrs1 gene. In contrast, Sharma et al. (2018) identified one GPE-linked SNPs for either Halle (Germany) or Dundee (Scotland), both of which reside 4 cM from Vrs1. Thus markers identified in this study should be much more useful and informative for subsequent fine mapping and marker-assisted selection for breeding to improve yields and yield components in barley.
For TGW: the TGW QTLs detected in this study overlapped with 11, 11, and 22 QTLs reported for TGW when the HEB-25 was evaluated at Halle (Germany), Dundee (Scotland) by Sharma et al. (2018), and Dubai (United Arab Emirates-UAE) by Saade et al. (2016), respectively. Notably, among four locations where HEB-25 was tested for TGW globally, TGW QTL, where the wild allele expressed the most significant positive effect, were usually environment-specific. For example, it was 3H-96.3 cM for Halle (Germany), 3H-107.8 cM for Dundee (Scotland), 6H-110.9 (Charlick, South Australia), and 3H-51.5 for Dubai (UAE). Thus, barley breeders should exercise caution depending on the target environment when selecting beneficial wild alleles from the HEB-25 population.
Among ten common TGW QTLs shared across the four global field trials with the HEB-25 in Germany, Scotland, UAE, and Australia, QTLs that reside closely to genes regulating flowering time (HvELF3, HvAP2(Zeo), HvPRR95, HvPRR1/HvTOC1/HvCO5/HvCO7) and row-type phenotype (Vrs1/3/4/5) were found to govern the TGW trait at all sites (Online Resource 3). The only common QTLs that increased TGW in all families at all four evaluating locations was QTgw.HEB25-5H.1 (1 cM). The wild alleles at this QTLS increased TGW in both control and salt-stressed field conditions and 2 years with the stark contrast in precipitation in Australia. Therefore, it could serve as a novel marker to add to the breeders’ toolbox to improve TGW in barley, especially in water-limiting or salt-stressed environments. This locus also merits further investigation as no known genes can be aligned to this locus thus far.
Conclusions
In this study, the field trial of the HEB-25 NAM population conducted in Australia identified QTLs unique to the Australian environment and different from the counterpart studies in the northern hemisphere. Our study showcased that genes known to regulate the flowering time and spike morphology in barley were pivotal in determining yield component traits like seed size and weight.
There were 18 hotspots associated with multiple grain size/weight traits in which one of them had wild alleles exerting a positive effect on both TGW and GPE. The wild alleles of genes/loci lying within this hotspot could serve as a valuable source for improving yield in Australia and worldwide.
Data availability
Supporting data and sources used for this manuscript are provided in Online Resource 1: Figure S1, Online Resource 2: Table S1, Online Resource 3: Table S2, Online Resource 4: Tables S3, Online Resource 5: Tables S4. Raw data generated in this work (approximately 2 Mbytes of data) are available from the corresponding author on reasonable request.
Code availability
Scripts for analysis in ASREML-R and SAS softwares were deposited in Figshare and available at https://doi.org/https://doi.org/10.25909/c.6286965.v1.
References
ABARES (2019) Agricultural commodities report. Coarse grains: March quarter 2019. Australian Bureau of Agricultural and Resource Economics and Sciences, Canberra, ACT. Available at: http://www.agriculture.gov.au/abares/research-topics/agricultural-commodities/mar-2019/coarse-grains (accessed 10 May 2019)
Barley. Available from : https://www.barleyaustralia.com.au/industry/barley/. Accessed May 17th 2022
Bayer MM, Rapazote-Flores P, Ganal M, Hedley PE, Macaulay M, Plieske J, Ramsay L, Russell J, Shaw PD, Thomas W, Waugh R (2017) Development and evaluation of a barley 50k iSelect SNP array. Front Plant Sci 8:1792
Bi X, Esse G, Mulki M, Kirschner G, Zhong J, Simon R, Korff M (2019) CENTRORADIALIS interacts with FLOWERING LOCUS T-like genes to control spikelet initiation, floret development and grain number. Plant Physiol 180:01454–02018. https://doi.org/10.1104/pp.18.01454
Brown AHD, Munday J, Oram RN (1988) Use of isozyme-marked segments from wild barley (Hordeum spontaneum) in barley breeding. Plant Breed 100:280–288. https://doi.org/10.1111/j.1439-0523.1988.tb00254.x
Bulman P, Mather DE, Smith DL (1993) Genetic improvement of spring barley cultivars grown in eastern Canada from 1910 to 1988. Euphytica 71:35–48
Butler DG, Cullis BR, Gilmour AR, Gogel BJ (2018) ASReml-R reference manual (Version 4). School of Mathematics and Applied Statistics, University of Wollongong
Büttner B, Draba V, Pillen K, Schweizer G, Maurer A (2020) Identification of QTLs conferring resistance to scald (Rhynchosporium commune) in the barley nested association mapping population HEB-25. BMC Genomics 21:837. https://doi.org/10.1186/s12864-020-07258-7
Cantalapiedra CP, Boudiar R, Casas AM, Igartua E, Contreras-Moreira B (2015) BARLEYMAP: physical and genetic mapping of nucleotide sequences and annotation of surrounding loci in barley. Mol Breed 35:13. https://doi.org/10.1007/s11032-015-0253-1
Capettini F, Ceccarelli S, Grando S (2010) Barley breeding history, progress, objectives, and technology. In: Ullrich SE (ed) Barley production, improvement, and uses. Wiley-Blackwell, Ames, IA, USA, pp 210–220
Chen C, Begcy K, Liu K, Folsom JJ, Wang Z, Zhang C, Walia H (2016) Heat stress yields a unique MADS box transcription factor in determining seed size and thermal sensitivity. Plant Physiol 171:606–622. https://doi.org/10.1104/pp.15.01992
Cu ST et al (2016) Genetic analysis of grain and malt quality in an elite barley population. Mol Breed 36:129. https://doi.org/10.1007/s11032-016-0554-z
Cullis BR, Smith AB, Coombes NE (2006) On the design of early generation variety trials with correlated data. J Agric Biol Environ Stat 11:381. https://doi.org/10.1198/108571106x154443
Dockter C et al (2014) Induced variations in brassinosteroid genes define barley height and sturdiness, and expand the green revolution genetic toolkit. Plant Physiol 166:1912–1927
Donald CM (1979) A barley breeding programme based on an ideotype. J Agric Sci 93:261–269. https://doi.org/10.1017/S0021859600037941
Eagles HA, Moody DB (2004) Using unbalanced data from a barley breeding program to estimate gene effects: the Ha2, Ha4, and sdw1 genes Australian. J Agric Res 55:379–387. https://doi.org/10.1071/AR03190
Finlay K, Wilkinson G (1963) The analysis of adaptation in a plant-breeding programme. Aust J Agric Res 14:742–754. https://doi.org/10.1071/AR9630742
Gage JL, Monier B, Giri A, Buckler ES (2020) Ten years of the maize nested association mapping population: impact, limitations, and future directions. Plant Cell 32:2083–2093. https://doi.org/10.1105/tpc.19.00951
Garrick DJ, Taylor JF, Fernando RL (2009) Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet Sel Evol 41:55
Griffiths S et al (2015) Genetic dissection of grain size and grain number trade-offs in CIMMYT wheat germplasm. PLoS ONE 10:e0118847. https://doi.org/10.1371/journal.pone.0118847
Gumedze FN, Welham SJ, Gogel BJ, Thompson R (2010) A variance shift model for detection of outliers in the linear mixed model. Comput Stat Data Anal 54:2128–2144
Hadjichristodoulou A (1990) Stability of 1000-grain weight and its relation with other traits of barley in dry areas. Euphytica 51:11–17. https://doi.org/10.1007/bf00022887
Hellewell K, Rasmusson D, Gallo-Meagher M (2000) Enhancing yield of semidwarf barley. Crop Sci. https://doi.org/10.2135/cropsci2000.402352x
Herzig P et al (2018) Contrasting genetic regulation of plant development in wild barley grown in two European environments revealed by nested association mapping. J Exp Bot 69:1517–1531. https://doi.org/10.1093/jxb/ery002
Hochman Z et al (2013) Reprint of “Quantifying yield gaps in rainfed cropping systems: a case study of wheat in Australia.” Field Crops Res 143:65–75. https://doi.org/10.1016/j.fcr.2013.02.001
Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6(2):65–70
Honsdorf N, March TJ, Pillen K (2017) QTL controlling grain filling under terminal drought stress in a set of wild barley introgression lines. PLOS ONE 12(10):e0185983
Houston K et al (2013) Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proc Natl Acad Sci 110:16675–16680. https://doi.org/10.1073/pnas.1311681110
Hu J et al (2018) Genetic properties of a nested association mapping population constructed with semi-winter and spring oilseed rapes. Front Plant Sci. https://doi.org/10.3389/fpls.2018.01740
Jedel PE, Helm JH (1994) Assessment of western Canadian barleys of historical interest: I. yield and agronomic traits. Crop Sci 34:922–927
Jia Q, Zhang J, Westcott S, Zhang X-Q, Bellgard M, Lance R, Li C (2009) GA-20 oxidase as a candidate for the semidwarf gene sdw1/denso in barley. Funct Integr Genom 9:255–262. https://doi.org/10.1007/s10142-009-0120-4
Jia Q et al (2016) Marker development using SLAF-seq and whole-genome shotgun strategy to fine-map the semi-dwarf gene ari-e in barley. BMC Genom 17:911. https://doi.org/10.1186/s12864-016-3247-4
Jiang QY, Hou J, Hao CY, Wang LF, Ge HM, Dong YS, Zhang XY (2011) The wheat (T. aestivum) sucrose synthase 2 gene (TaSus2) active in endosperm development is associated with yield traits. Funct Integr Genom 11:49–61. https://doi.org/10.1007/s10142-010-0188-x
Jiang YM et al (2015) A yield-associated gene TaCWI, in wheat: its function, selection and evolution in global breeding revealed by haplotype analysis. Theor Appl Genet 128:131–143. https://doi.org/10.1007/s00122-014-2417-5
Kalladan R et al (2013) Identification of quantitative trait loci contributing to yield and seed quality parameters under terminal drought in barley advanced backcross lines. Mol Breed 32:71–90
Kitony JK et al (2021) Development of an Aus-derived nested association mapping (Aus-NAM) population in rice. Plants (basel, Switzerland). https://doi.org/10.3390/plants10061255
Komatsuda T et al (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA 104:1424–1429. https://doi.org/10.1073/pnas.0608580104
Koppolu R et al (2013) Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc Natl Acad Sci USA 110:13198–13203. https://doi.org/10.1073/pnas.1221950110
Kurakawa T et al (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445:652–655. https://doi.org/10.1038/nature05504
Kuroha T et al (2009) Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21:3152–3169. https://doi.org/10.1105/tpc.109.068676
Lakew B, Henry RJ, Ceccarelli S, Grando S, Eglinton J, Baum M (2013) Genetic analysis and phenotypic associations for drought tolerance in Hordeum spontaneum introgression lines using SSR and SNP markers. Euphytica 189:9–29
Langridge P, Lagudah ES, Holton TA, Appels R, Sharp PJ, Chalmers KJ (2001) Trends in genetic and genome analyses in wheat: a review. Aust J Agric Res 52:1043–1077. https://doi.org/10.1071/AR01082
Li JZ, Huang XQ, Heinrichs F, Ganal MW, Röder MS (2006) Analysis of QTLs for yield components, agronomic traits, and disease resistance in an advanced backcross population of spring barley. Genome 49:454–466. https://doi.org/10.1139/g05-128
Liu W et al (2011) Association mapping in an elite maize breeding population. Theor Appl Genet 123:847–858
Loscos J, Igartua E, Contreras-Moreira B, Gracia MP, Casas AM (2014) HvFT1 polymorphism and effect—survey of barley germplasm and expression analysis. Front Plant Sci. https://doi.org/10.3389/fpls.2014.00251
Lundqvist U (1997) New and revised descriptions of barley genes. Barley Genet Newslett 26:22–516
Luo J, Wei B, Han J, Liao Y, Liu Y (2019) Spermidine increases the sucrose content in inferior grain of wheat and thereby promotes its grain filling. Front Plant Sci. https://doi.org/10.3389/fpls.2019.01309
Markova RN, Valcheva D, Valchev D, Mihajlov L, Karov I, Ilieva V (2015) Correlation between grain yield and yield components in winter barley varieties. Agric Sci Technol 7(1):40–44
Maurer A et al (2015) Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genom 16:290
Maurer A, Draba V, Pillen K (2016) Genomic dissection of plant development and its impact on thousand grain weight in barley through nested association mapping. J Exp Bot 67:2507–2518. https://doi.org/10.1093/jxb/erw070
Maurer A, Sannemann W, Léon J, Pillen K (2017) Estimating parent-specific QTL effects through cumulating linked identity-by-state SNP effects in multiparental populations. Heredity 118:477–485
Mickelson HR, Rasmusson DC (1994) Genes for short stature in barley. Crop Sci 34:1180–1183
Mikołajczak K et al (2016) Quantitative trait loci for yield and yield-related traits in spring barley populations derived from crosses between European and Syrian cultivars. PLoS ONE 11:e0155938. https://doi.org/10.1371/journal.pone.0155938
Nadolska-Orczyk A, Rajchel IK, Orczyk W, Gasparis S (2017) Major genes determining yield-related traits in wheat and barley. Theor Appl Genet 130:1081–1098. https://doi.org/10.1007/s00122-017-2880-x
Nevo E, Shewry P (1992) Origin, evolution, population genetics and resources for breeding of wild barley, Hordeum spontaneum, in the fertile crescent. In: Shewry PR (ed) Genetics, biochemistry, molecular biology and biotechnology. C.A.B. International, The Alden Press, Oxford, pp 19–43
Patterson HD, Thompson R (1971) Recovery of inter-block information when block sizes are unequal. Biometrika 58:545–554. https://doi.org/10.2307/2334389
Paul MJ, Watson A, Griffiths CA (2020) Trehalose 6-phosphate signalling and impact on crop yield. Biochem Soc Trans 48:2127–2137. https://doi.org/10.1042/bst20200286
Peltonen-Sainio P, Kangas A, Salo Y, Jauhiainen L (2007) Grain number dominates grain weight in temperate cereal yield determination: evidence based on 30 years of multi-location trials. Field Crops Res 100:179–188. https://doi.org/10.1016/j.fcr.2006.07.002
Pham A-T et al (2019) Genome-wide association of barley plant growth under drought stress using a nested association mapping population. BMC Plant Biol 19:134. https://doi.org/10.1186/s12870-019-1723-0
Pham A-T, Maurer A, Pillen K, Taylor J, Coventry S, Eglinton JK, March TJ (2020) Identification of wild barley derived alleles associated with plant development in an Australian environment. Euphytica 216:148. https://doi.org/10.1007/s10681-020-02686-8
Pu-yang D et al (2022) A major and stable QTL for wheat spikelet number per spike validated in different genetic backgrounds. J Integr Agric 21:1551–1562. https://doi.org/10.1016/S2095-3119(20)63602-4
Radchuk V et al (2021) Grain filling in barley relies on developmentally controlled programmed cell death. Commun Biol 4:428. https://doi.org/10.1038/s42003-021-01953-1
Ramsay L et al. (2011) INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nature genetics 43:169
Ren X, Sun D, Sun G, Li C, Dong W (2013) Molecular detection of QTL for agronomic and quality traits in a doubled haploid barley population. Aust J Crop Sci 7(6):878–886
Richards RA (1991) Crop improvement for temperate Australia: future opportunities. Field Crop Res 26:141–169. https://doi.org/10.1016/0378-4290(91)90033-R
Saade S et al (2016) Yield-related salinity tolerance traits identified in a nested association mapping (NAM) population of wild barley. Sci Rep 6:32586. https://doi.org/10.1038/srep32586
Sadras VO, Rebetzke GJ (2013) Plasticity of wheat grain yield is associated with plasticity of ear number. Crop Pasture Sci 64:234–243. https://doi.org/10.1071/CP13117
Schils R et al (2018) Cereal yield gaps across Europe. Eur J Agron 101:109–120. https://doi.org/10.1016/j.eja.2018.09.003
Schmalenbach I, March TJ, Bringezu T, Waugh R, Pillen K (2011) High-resolution genotyping of wild barley introgression lines and fine-mapping of the threshability locus thresh-1 using the illumina goldengate assay. G3 (Bethesda, Md) 1: 187–196. https://doi.org/10.1534/g3.111.000182
Sharma R et al (2018) Genome-wide association of yield traits in a nested association mapping population of barley reveals new gene diversity for future breeding. J Exp Bot 69:3811–3822. https://doi.org/10.1093/jxb/ery178
Slafer GA (2003) Genetic basis of yield as viewed from a crop physiologist’s perspective. Ann Appl Biol 142:117–128. https://doi.org/10.1111/j.1744-7348.2003.tb00237.x
Spragg J (2016) Australian feed grain supply and demand report 2016 JCS Solutions Pty Ltd: North Victoria, Australia:1–42
Tokunaga H, Kojima M, Kuroha T, Ishida T, Sugimoto K, Kiba T, Sakakibara H (2012) Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. The Plant J 69:355–365. https://doi.org/10.1111/j.1365-313X.2011.04795.x
Van Gool D, Vernon L (2006) Potential impacts of climate change on agricultural land use suitability: Barley. Department of Primary Industries and Regional Development, Western Australia, Perth Report 302
Vatter T, Maurer A, Kopahnke D, Perovic D, Ordon F, Pillen K (2017) A nested association mapping population identifies multiple small effect QTL conferring resistance against net blotch (Pyrenophora teres f. teres) in wild barley. PLoS ONE 12:e0186803. https://doi.org/10.1371/journal.pone.0186803
Vatter T, Maurer A, Perovic D, Kopahnke D, Pillen K, Ordon F (2018) Identification of QTL conferring resistance to stripe rust (Puccinia striiformis f. sp. hordei) and leaf rust (Puccinia hordei) in barley using nested association mapping (NAM). PLoS ONE 13:e0191666. https://doi.org/10.1371/journal.pone.0191666
Walker CK, Ford R, Muñoz-Amatriaín M, Panozzo JF (2013) The detection of QTLs in barley associated with endosperm hardness, grain density, grain size and malting quality using rapid phenotyping tools. Theor Appl Genet 126:2533–2551. https://doi.org/10.1007/s00122-013-2153-2
Wang G, Schmalenbach I, von Korff M, Léon J, Kilian B, Rode J, Pillen K (2010a) Association of barley photoperiod and vernalization genes with QTLs for flowering time and agronomic traits in a BC2DH population and a set of wild barley introgression lines. Theor Appl Genet 120:1559–1574. https://doi.org/10.1007/s00122-010-1276-y
Wang J, Yang J, McNeil DL, Zhou M (2010b) Identification and molecular mapping of a dwarfing gene in barley (Hordeum vulgare L.) and its correlation with other agronomic traits. Euphytica 175:331–342
Wang J, Sun G, Ren X, Li C, Liu L, Wang Q, et al (2016) QTL underlying some agronomic traits in barley detected by SNP markers. BMC Genet 17(1):103
Wang Q et al (2019) Dissecting the genetic basis of grain size and weight in barley (Hordeum vulgare L.) by QTL and comparative genetic analyses. Front Plant Sci. https://doi.org/10.3389/fpls.2019.00469
Wendler N, Mascher M, Nöh C, Himmelbach A, Scholz U, Ruge-Wehling B, Stein N (2014) Unlocking the secondary gene-pool of barley with next-generation sequencing. Plant Biotechnol J 12:1122–1131. https://doi.org/10.1111/pbi.12219
Whan AP, Smith AB, Cavanagh CR, Ral J-PF, Shaw LM, Howitt CA, Bischof L (2014) GrainScan: a low cost, fast method for grain size and colour measurements. Plant Methods 10:23. https://doi.org/10.1186/1746-4811-10-23
Wiegmann M et al (2019) Barley yield formation under abiotic stress depends on the interplay between flowering time genes and environmental cues. Sci Rep 9:6397. https://doi.org/10.1038/s41598-019-42673-1
Wuriyanghan H et al (2009) The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell 21:1473–1494. https://doi.org/10.1105/tpc.108.065391
Xu X et al (2018) Genome-wide association analysis of grain yield-associated traits in a Pan-European barley cultivar collection. The Plant Genome. https://doi.org/10.3835/plantgenome2017.08.0073
Yang X et al (2012) Live and let die-the Bsister MADS-box gene OsMADS29 controls the degeneration of cells in maternal tissues during seed development of rice (Oryza sativa). PLoS ONE 7:e51435. https://doi.org/10.1371/journal.pone.0051435
Zalewski W, Galuszka P, Gasparis S, Orczyk W, Nadolska-Orczyk A (2010) Silencing of the HvCKX1 gene decreases the cytokinin oxidase/dehydrogenase level in barley and leads to higher plant productivity. J Exp Bot 61:1839–1851. https://doi.org/10.1093/jxb/erq052
Zalewski W, Orczyk W, Gasparis S, Nadolska-Orczyk A (2012) HvCKX2 gene silencing by biolistic or Agrobacterium-mediated transformation in barley leads to different phenotypes. BMC Plant Biol 12:206
Zalewski W, Gasparis S, Boczkowska M, Rajchel IK, Kała M, Orczyk W, Nadolska-Orczyk A (2014) Expression patterns of HvCKX genes indicate their role in growth and reproductive development of barley. PLoS ONE 9:e115729
Zhang Y, Sun Y, Sun J, Feng H, Wang Y (2019) Identification and validation of major and minor QTLs controlling seed coat color in Brassica Rapa L. Breed Sci 69:47–54. https://doi.org/10.1270/jsbbs.18108
Zhou MX (2010) Barley production and consumption. In: Zhang G, Li C (eds) Genetics and improvement of barley malt quality. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 1–17. https://doi.org/10.1007/978-3-642-01279-2_1
Zhou H et al (2016) Mapping and validation of major quantitative trait loci for kernel length in wild barley (Hordeum vulgare ssp. spontaneum). BMC Genet 17:130. https://doi.org/10.1186/s12863-016-0438-6
Acknowledgements
The authors would like to thank Giang Truong Nguyen, Hoa Thanh Thi Nguyen, Simon Schreiber, and Pauline Delano for their technical assistance with measuring and phenotyping in both field and laboratory conditions.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This study was funded by the Grains Research and Development Corporation (project UA00148).
Author information
Authors and Affiliations
Contributions
A-TP, SC, JE, TM, TDN carried out the experiments. A-TP, AM and JT analysed the data. KP provided seed material. JE, SC and TM provided crucial information for the design of the experiments. A-TP and TM wrote the manuscript. All authors read, revised and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10681_2023_3260_MOESM1_ESM.png
Fig. S1.Bar plots showing trait differences between the HEB-25 population and 11 barley check varieties sown in the 2015 (A) and 2016 (B)’s experiments. (PNG 222 KB)
10681_2023_3260_MOESM2_ESM.xlsx
Table S1. The monthly and annual rainfalls for 2015, 2016, and 26-year-average recorded at Charlick, Strathalbyn, South Australia (XLSX 10 KB)
10681_2023_3260_MOESM3_ESM.xlsx
Table S2. Overview of all QTL and their significant effects on seven traits studied in 25 families of the HEB-25 population (XLSX 113 KB)
10681_2023_3260_MOESM5_ESM.xlsx
Table S4. List of genes residing 4 cM upstream and downstream of the markers within three beneficial hotspots (XLSX 20 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pham, AT., Maurer, A., Pillen, K. et al. A wild barley nested association mapping population shows a wide variation for yield-associated traits to be used for breeding in Australian environment. Euphytica 220, 24 (2024). https://doi.org/10.1007/s10681-023-03260-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-023-03260-8