Abstract
Many in situ conservation programs have been developed to preserve plant landrace diversity and to promote its sustainable utilization, but little is known about the effectiveness of the developed programs in conserving plant genetic diversity. We investigated the effectiveness of an unregulated (i.e., unplanned or open) conservation system maintained by Thai farmers in conserving Thai elite cassava (Manihot esculenta Crantz) varieties. Specifically, we compared genetic diversity of 266 cassava clones that were collected from 80 farms in eight provinces with 16 cassava landraces and varieties released since the 1970s through genotyping with 35 informative simple sequence repeat (SSR) markers. The SSR analysis revealed a large regional heterogeneity in cassava diversity, with a strong genetic differentiation of the assayed clones among the 80 farms (19.8 %) and across the eight provinces (11.8 %). Significant associations were also found between SSR variation and farm agro-ecological factors or some farming practices. However, there was no significant genetic differentiation (0.9 %) between the 266 farm clones and 16 reference varieties. These findings suggest that the Thai elite cassava genetic diversity was fortuitously conserved by the farmers through farming with different sets of varieties. Implications of these findings are discussed with respect to on-farm conservation of plant genetic resources.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recent decades have seen an increasing role of in situ conservation (i.e., the conservation in its natural habitat) of plant genetic resources (Maxted et al. 1997; Jarvis et al. 2007; Padulosi et al. 2012), and many in situ and on-farm conservation programs have been developed to preserve plant landrace diversity and to promote its sustainable utilization (e.g., see Veteläinen et al. 2009). These efforts reflect the recognition of on-farm conservation as an essential component of sustainable agriculture and the appreciation of traditional farmers in managing crop genetic diversity (Altieri and Merrick 1987; Brush 1991, 2000; CBD 2002). However, how effective these newly developed programs are in conserving plant genetic diversity remains to be determined (Jarvis et al. 2008; Padulosi et al. 2012; Nevo et al. 2012; Thomas et al. 2012). Much could be learnt for the development of effective conservation programs from unregulated (i.e., unplanned or open) conservation systems such as traditional agroecosystems (e.g., Duputié et al. 2009; McKey et al. 2010), home gardens (e.g., Rocha et al. 2008), and unprotected natural ecosystems (e.g., Nevo et al. 2012). This paper examines the effectiveness of an unregulated conservation system maintained by the Thai farmers in conserving Thai elite cassava (Manihot esculenta Crantz) varieties, with the hope it will provide useful data for informing current in situ conservation efforts.
Cassava is one of the world’s most important tropical crop plants (Cock 1985; FAO/IFAD 2001) and the fourth most important food calorie crop in the tropics, and has been growing in importance both for food security (especially in Africa) and for multiple commercial and industrial uses (mainly in Latin America and Asia) (Debouck et al. 2011). This crop has been a model plant for the studies of the evolutionary ecology of clonally propagated domesticated plants (e.g., see McKey et al. 2010) and the maintenance of genetic diversity in traditional agroecosystems (e.g., see Salick et al. 1997; Sambatti et al. 2001; Elias et al. 2004; Turyagyenda et al. 2012). Most of the diversity studies were performed using simple sequence repeat (SSR) markers (Fregene et al. 2003; Montero-Rojas et al. 2011) and largely in the region of cassava domestication in South America (e.g. Elias et al. 2004; Montero-Rojas et al. 2011). Some research outcomes were encouraging, helping advance our understanding about diversity maintenance and its relevance to in situ conservation (e.g., Elias et al. 2000, 2001; Sambatti et al. 2001; Duputié et al. 2009; Siqueira et al. 2009; Montero-Rojas et al. 2011). For example, traditional cassava-farming systems have been shown to maintain genetic diversity by means of recombination, gene flow and somatic mutation (see Elias et al. 2000, 2001; Sambatti et al. 2001) and large cassava diversity was found to be conserved in home gardens (see Rocha et al. 2008).
Cassava is one of the most important economic crops in Thailand with 80 % of the fresh root production destined for export to the European Union and Asian countries (Ratanawaraha et al. 2001). Thai cassava sector was re-energized when it capitalized on European market opportunities for dried chips and pellets, beginning in the 1970s (Hershey and Debouck 2010). In 2012, 3.3 million acres of cassava were planted and 27 million tons of fresh cassava root were produced from 0.48 million farmers across 50 provinces (Office of Agricultural Economics 2013; ASEAN Economic Community 2013). Early breeding efforts since 1937 have contributed to the success of cassava production with the introduction of 20 varieties from Malaysia and the Philippines in the 1930s and 65 varieties from the Columbia and Virgin Islands from 1963 to 1977 (Rojanaridpiched et al. 2007). However, the cassava production has greatly increased only after the release of the first Thai variety Rayong 1 in 1975. Rayong 1 replaced the early introductions and has been dominant in cassava production during 1970–1990s before being replaced by Kasetsart 50 released in 1992 with higher yield and better adaptation to unfavorable conditions (Sarakarn et al. 2001). So far, the Thai cassava breeding has officially released 13 bitter-type and one sweet-type cassava varieties (Sarakarn et al. 2007; Department of Agriculture 2013).
A companion study which aimed to identify genetically distinct cassava clones from Thai cassava farming (Wangsomnuk et al. 2013) revealed a diversity of distinct cassava clones present on different farms. An initial farm variety use survey showed that half of the Thai cassava varieties released since the 1970s was still cultivated and most of the farms planted with only one to three varieties (supplementary Table S1). However, not all of the surveyed farms had accurate records of variety acquisition, plantation and maintenance. These observations inspired us to investigate further whether and how genetic diversity of the Thai cassava varieties released since the 1970s was conserved on farms. Based on the findings from the companion study (Wangsomnuk et al. 2013), we formulated a working hypothesis that the Thai elite cassava genetic diversity may have been un-intentionally conserved by the farmers in response to industrial demands on cassava.
During the investigation, we realized that Thai cassava farming may represent a unique and unregulated conservation system of plant genetic resources maintained by farmers, similar to those operating in home gardens of Costa Rica (Rocha et al. 2008). First, no specific efforts have been documented to develop an official on-farm conservation program for local, adapted cassava germplasm (FAO 2010). This may reflect the fact that cassava varieties are largely clonally propagated; there are few unique landrace varieties in Thailand (Hershey and Debouck 2010); and most importantly, the official varieties released since the 1970s are conserved in the Thai cassava collection. Second, Thai agricultural farming has been modernized since 1960s and may differ from those traditional cassava farming systems in South America in variety acquisition, plantation, and maintenance (Ratanawaraha et al. 2001), as the transition has been made for Thai cassava from a staple food to products and raw materials for the processing industry (FAO/IFAD 2001) and the adoption rate of new varieties by farmers is high (Debouck et al. 2011). The Thai cassava farming is relatively young, with insufficient generations for mutation and natural selection forces to have played a major role, and may be sensitive to diversity assessment, without a large input of new varieties since the 1970s. These unique features, along with the ex situ conservation of the Thai elite cassava varieties, should provide an opportunity to assess the unregulated on-farm conservation of Thai cassava germplasm.
The specific objectives of this investigation were to (1) compare the genetic diversity and structure of 266 cassava clones of unknown genetic background that were collected from 80 farms in eight provinces with 16 Thai cassava landraces and varieties released since the 1970s through genotyping with 35 informative SSR markers, (2) identify varieties within the farm clones and evaluate their on-farm distributions across the provinces, and (3) determine the associations between on-farm SSR variation and farm geographic location, agroecological environment, farming practice, or farm production history. As the direct assessment of on-farm variety distribution is not feasible, we considered the alternative assessment of the genetic associations between genetic variation within farm clones and the features of farming systems. Any significant genetic association would signal an uneven distribution of varieties on farms, since such a genetic association cannot be established theoretically from a uniform on-farm variety distribution across the provinces.
Materials and methods
Plant materials
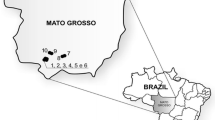
The cassava samples studied here consisted of 266 clones of unknown genetic background that were collected from 80 farms each with a cassava planting area of 5,000–32,000 m2 and 16 Thai landraces and varieties released since the 1970s as reference varieties for diversity comparison (Table S1). The surveyed farms were located in 16 districts and eight provinces (Fig. 1 and Table 1) and were selected to represent major cassava planting areas in northeast, north, east, and west Thailand. On farm collection of cassava clones was conducted from November 2011 to April 2012. Five clones per farm were selected based only on our observations of phenotypic variation. The farm locality data (i.e., latitude, longitude, altitude) was also obtained. The collected stems were re-planted in Nong Lek Subdistrict, Kosum Phisai District, Maha Sarakham province for further phenotypic and genetic characterization. The 16 reference varieties were sampled from Rayong Field Crops Research Center and Khon Kaen Field Crops Research Center, Thailand and represented two cassava types (bitter and sweet). Thirteen bitter cassava varieties had been officially released since the 1970s from several Thai breeding programs (Ratanawaraha et al. 2001; Sarakarn et al. 2007; Department of Agriculture 2013). Rayong 2 was a sweet cassava variety released in 1984 (Sarakarn et al. 2007). Two sweet cassava landraces Hanatee and Munsuan were cultivated before 1984 (Rojanaridpiched 1988).
Geographic locations of 80 studied farms covering eight provinces in Thailand. A unique survey number (1–80) was attributed to each farm listed in Table 1 and forms part of a farm name for identification. The farms are colored for different provinces and their soil series are also shown (see Table S5 for the soil series labeling). (Color figure online)
To facilitate genetic diversity analyses, the same farms were re-surveyed in October, 2012, to collect further information on farm age and plantation area; cassava planting history, area and production; and farm use of other crops, fertilizers and hormones. To analyze the associations between genetic diversity and agroecological environment or climate factors, the soil data at the farm level were obtained (Office of Soil Resources Survey and Research 2011a, b; Office of Soil Survey and Land Use Planning 2004a, b, c). Annual rainfall, temperature and humidity data at the provincial level were obtained from the inventory files dated from August 2011 to July 2012 from Thai Meteorological Department (2012).
DNA extraction and SSR analysis
The genomic DNA extraction was performed for the 16 reference varieties based on a pool of young leaves from three individual plants per variety and for the 400 farm clones based on the young leaf tissue from each plant collected in the field. The detailed procedures used for DNA extraction and SSR analysis were described in the companion study on clone identification for cassava breeding (see Wangsomnuk et al. 2013). Briefly, 24 genomic SSR and 17 EST-derived SSR markers were selected from published literature based on marker type, informativeness and linkage group (Chavarriaga-Aguirre et al. 1998; Mba et al. 2001; Raji et al. 2009; Kunkeaw et al. 2010) for the SSR analysis (Table S2). An initial screening of the 400 farm clones was performed with three genomic SSR and three EST-SSR markers to identify and remove clone duplications from each farm. Based on the SSR profiles, 266 putatively distinct clones for all 80 farms (Table 1) were identified following the method recommended by Arnaud-Haond et al. (2007) and used, along with the 16 reference varieties, for final genotyping with the 41 SSR markers.
Data analysis
SSR data were analyzed for the level of polymorphism with respect to primer and sample origin (i.e., farm, district and province) by counting the number of polymorphic alleles and generating summary statistics of allelic frequencies. The numbers of alleles detected by all primer pairs were plotted against their frequencies of occurrence in all assayed samples. As cassava ploidy is uncertain (i.e., either diploid or allotetraploid with disomic inheritance; Awoleye et al. 1994; Jennings 1995; Raji et al. 2009) and only two of the 41 SSR markers appeared to display a di-allelic profile, Shannon’s entropy was calculated following Reyes-Valdes and Williams (2005) to estimate the diversity content per locus, as this estimate does not require strict genetic assumptions such as marker inheritance and sample ploidy. The entropy-based diversity content (eDC) provides a measure of the effective number of alleles per marker locus (Reyes-Valdes and Williams 2005). To assess group-specific allelic richness, the eDC per sample was also estimated for cassava groups of variable sample size. The estimates of the group-specific eDC per sample were compared for allelic richness among various groups of the assayed samples with respect to sample origin and other factors such as cassava type, variety and other crop use, fertilizer and hormone use. The estimations of eDC were made by using a SAS program written in SAS IML (SAS Institute Inc. 2008) which is available upon request from the first author. Additional allelic counts were made for various groups of the assayed samples and allelic differences among groups of variable sample size were tested for significance following the random permutation method of Fu (2010).
The analysis of molecular variance (AMOVA) was performed using Arlequin version 3.5 (Excoffier and Lischer 2010) to assess genetic diversity and structure of the assayed samples. The cassava samples were grouped based on sample origin and characteristics associated with the surveyed farms. The prior defined groups considered here were: 80 farms versus 16 varieties, 80 farms, 16 districts, 8 provinces, bitter versus sweet cassava types and 21 soil types. The Arlequin program provides a partitioning of the total SSR variation into within- and among-group components and allows for a measuring of inter-group distances as a proportion of the total SSR variation residing between any two groups (Phi statistic) (Excoffier et al. 1992). Significance of resulting variance components and inter-group genetic distances was tested with 10,000 random permutations.
The structural inference for the prior defined groups was specifically done by estimating genetic distances among groups and clustering the prior defined groups. The neighbor joining (NJ) clustering of the prior defined groups was made using NTSYS-PC 2.01 (Rohlf 1997) based on pairwise estimates of genetic distance from AMOVA for each structural model. A distogram based on the AMOVA estimates of genetic distances was plotted using R package “squash” (Eklund 2011; R Development Core Team 2013).
The genetic structure was also inferred without considering prior information and the inference was done using the program STRUCTURE version 2.2.3 (Pritchard et al. 2000; Falush et al. 2007). The STRUCTURE program was run 40 times for each subpopulation (K) value, ranging from 2 to 15, using the admixture model with 10,000 replicates for burn-in and 10,000 replicates during analysis. The final population subgroups were determined based on (1) likelihood plot of these models, (2) the change in the second derivative (∆K) of the relationship between K and the log-likelihood (Evanno et al. 2005), and (3) stability of grouping patterns across 40 runs. For a given K with 40 runs, the run with the highest likelihood value was selected to assign the posterior membership coefficients to each sample. A graphical bar plot was then generated with the posterior membership coefficients. The size and composition of each optimal cluster with respect to farm, district and province were analyzed. Additional AMOVA was also made to quantify genetic differentiation among the inferred optimal clusters.
The inferred genetic structure was further compared for consistency with the genetic relationships of individual samples. An NJ analysis of the 282 samples was also made using PAUP* (Swofford 1998) based on the original data of 365 SSR alleles and a radiation tree was displayed using MEGA 3.01 (Kumar et al. 2004). A principal coordinate analysis (PCoA) of the 282 cassava samples was performed using NTSYS-PC 2.01 (Rohlf 1997) based on the similarity matrix of 365 SSR alleles, and plots of the first three resulting principal components were made to assess the sample associations.
To assess associations between on-farm SSR variation and farming features, additional AMOVA was performed to test and quantify genetic differentiation among the farm clones associated with the categorical farming variables such as the soil type, the number of historical cassava varieties and other crops planted, the number of different fertilizers used, and the use of hormones or not. To assess the associations between detected SSR variation and quantitative variables such as farm geographic location, size, age and climate data, Mantel tests were conducted using GenAlEx v6 software (Peakall and Smouse 2006) with and without consideration of isolation by distance model (Rousset1997). A correlation analysis was also made for all the assessed farm variables using SAS PROC CORR (SAS Institute Inc 2008) to generate the Pearson correlation matrix.
The variety identification was performed from the 266 farm clones based on the presence of a SSR allele or a combination of two SSR alleles unique to a given variety; that is, the allele or combination of two alleles was found to be present only in one of the 16 reference varieties. The identified farm clones were grouped according to their farm origins with respect to province to infer the evenness of variety distribution in the Thai cassava production region. Similarly, the distribution evenness was also assessed for the 11 farm clone clusters that were inferred using STRUCTURE.
Results
SSR variation
The SSR analysis revealed that six of the 17 EST-derived SSR markers displayed only monomorphic bands for all 282 samples and thus they were removed from further analyses. The other 35 markers revealed two monomorphic and 365 polymorphic alleles in the 282 samples (Table S2) and only polymorphic allele data were used for genetic diversity analyses. The number of alleles detected per locus ranged from 2 to 21 and averaged 10.4. The observed frequency of an individual allele ranged from 0.028 to 0.996 with an average of 0.597. There were 18 alleles observed for every 0.05 interval of allelic spectrum, seven alleles of frequency smaller than 0.05, and 55 alleles of frequency larger than 0.95 (supplementary Fig. S1). Interestingly, 11 EST-derived SSR markers detected only 61 alleles, while the 24 genomic SSR primer pairs identified 304 alleles. The most informative primer pair was the genomic SSRY235 on linkage group G with an eDC value of 5.27 and 18 alleles detected, followed by the genomic GA5 on linkage group Q with an eDC value of 4.93 and 21 alleles detected (Table S2). These 35 SSR markers sampled SSR alleles in either transcribed or non-transcribed chromosomal regions and thus should provide an adequate measure of genetic diversity.
Genetic diversity
The assayed cassava samples displayed different patterns of SSR variation with respect to variety, farm, district, province, and cassava type (i.e., bitter/sweet; Table 2 and Table S3). The AMOVA revealed a non-significant (p = 0.14) 0.88 % of the total SSR variance between the 16 reference varieties and 266 farm clones. The cassava samples among farms explained the largest SSR variance (19.8 %), followed by those among districts (13.3 %) and among provinces (11.8 %). Based on the estimates of group-specific entropy-based diversity content (SeDC) per sample, five farms with the highest SeDC per sample (>0.328) were NNN22, NNN21, KML55, SKK16 and SKC11, while six farms with the least SeDC per sample (<0.286) were SSK19, PPN61, MBB5, PKN69, KMT31 and MWD7 (Table 1). Two districts with the most SeDC per sample (0.312) were Sai Ngam and Mueang Kanchanaburi and two districts with the least SeDC per sample (<0.303) were Mancha Khiri and Watthana Nakhon (Table S4). Two provinces with the most SeDC per sample (0.308) were Kamphaeng Phet and Kanchanaburi, while Prachin Buri displayed the least SeDC per sample (0.300) (Table S4). Differentiation between bitter and sweet cassava also explained 5.4 % SSR variation (p < 0.001), with the bitter clones harboring more diversity per sample (SeDC = 0.307) than the sweet clones (SeDC = 0.291) (Table S4).
There were 41 (51 %) farms, 16 districts, and 8 provinces with cassava clones displaying significant genetic differentiations of variable magnitude from the reference varieties (Fig. S2). The farm, district, and province with cassava clones showing the largest genetic differentiations from the reference varieties were KSM43, Mueng Kamphaeng Phet, and Kamphaeng Phet, with pairwise SSR difference estimates of 0.202, 0.140 and 0.135, respectively. Without considering the reference varieties, significant genetic differentiation was also observed among the clones representing farms, districts and provinces (Fig. S2). For example, the clones from Maha Sarakham province showed the largest differentiation from those from Prachin Buri province (Table 3; Fig. S2). Similarly, the clones from Wapi Pathum district displayed the largest differentiation from those from Kabin Buri district.
Allelic count for the cassava samples with respect to variety, farm, district, province, and cassava type revealed patterns of variation (Tables 3 and S4) similar to those based on group-specific eDC per sample. For example, the 266 farm clones had the same number of SSR alleles (365) as the 16 reference varieties, but when the variable sample size was considered, the 16 reference varieties would be statistically expected to have only 356.1(±2.4) alleles and the 266 farm clones would expect to have 365.0(±0.4) alleles. Thus, the observed 365 alleles for the 16 reference varieties were more than expected (Table S4). There was a large number of significant cases of pairwise allelic differences for various groups of the assayed clones (Table S4). For example, a significant allelic difference of 12 was observed between the clones from Maha Sarakham and those from Si Sa Ket.
Genetic structure
The neighbor joining analysis of pairwise genetic distances detected seven major genetic clusters at the farm level (C1–C7; Fig. 2). These clusters displayed varied regional heterogeneity. While the clones representing farms from Si Sa Ket or Maha Sarakham province were largely clustered in Cluster 1 or 7, respectively, some farm clones were widely spread into various clusters regardless of their geographic provenance. For example, the clones for six farms from Prachin Buri were clustered in Cluster 6, but the clones for the other four farms from this province were located in Clusters 4, 5, and 7. The clustering at the district level was less variable than those at the farm level and displayed two major clusters (Fig. S3A). These two major clusters mirrored well those detected at the province level (Fig. S3B). The clones from the provinces of Maha Sarakham, Si Sa Ket, Nakhon Ratchasima and Khon Kaen formed one cluster (Fig. S3B), while the clones from the other four provinces were clustered together.
Genetic structure of 266 cassava clones representing 80 farms in Thailand as illustrated in the circular neighbor-joining tree based on pairwise genetic distances estimated from AMOVA. The farms representing provinces (see Table 1 for farm labeling) are highlighted in different colors corresponding to the colored province map of Thailand. Seven major genetic clusters (C1–C7) display varied regional heterogeneity. (Color figure online)
The prior defined structures described above accord well with the inferred genetic associations of individual clones (Fig. 3). The NJ tree revealed that more than six groups could be identified (Fig. 3A), while the PCoA plot showed two major groups (Fig. 3B) which were consistent with the genetic structure inferred at the province level. These association analyses also revealed the wide distributions of the 16 reference varieties over the PCoA plot and NJ tree (Fig. 3) and the narrow base of the 12 sweet cassava clones (results not shown). There were no groups exclusive to any province or district, as the clones from different provinces or districts were largely mixed into various groups (Fig. 3A).
Genetic relationships of 266 cassava clones collected from 80 farms and 16 reference varieties, as revealed in the neighbor-joining (NJ) tree (A) and PCoA plot (B). Farm clones and reference varieties are highlighted with filled and open circles, respectively. Farm clones representing provinces are highlighted in different colors corresponding to the colored province map of Thailand. The reference varieties were widely scattered over the NJ tree or PCoA plot. (Color figure online)
The STRUCTURE analysis revealed 11 optimal clusters in the 282 samples (Fig. 4A) with the log-likelihood of −33,393 (Fig. S4A) and the largest partition (25.2 %) of the total SSR variation (Table S3). The log-likelihood profile for the optimal clusters gained further support from the rate of change in the second derivative of the log-likelihoods over various Ks analyzed (Fig. S4B). The cluster size ranged from 12 (Cluster 5) to 44 (Cluster 11) and all the samples had multiple memberships across the 11 clusters (Fig. 4B). The largest cluster included varieties Rayong 2 and Rayong 7 and had clones from 27 farms, 11 districts and 7 provinces. In this largest cluster, four farms (KMS47, KMS48, KMS49 and KSS60) had three clones each, and the district and province with the largest number of clones were Mueang Kamphaeng Phet and Sa Kaeo, respectively. Interestingly, Cluster 8 with the dominant variety Kasetsart 50 had 27 clones from 23 farms, 11 districts and 8 provinces. Cluster 10 seems to consist dominantly of sweet cassava clones with two sweet cassava landraces Hanatee and Munsuan. The cluster composition with respect to farm was given in Table 1. Interestingly, 73 farms had clones present in more than one cluster, six farms in four clusters and one farm in five clusters.
Genetic structure of 282 cassava clones and varieties inferred using STRUCTURE for K = 10, 11, and 12 (A) and composition of 11 optimal clusters (B). Variety label is given in Table S1. FHNC stands for the farm with the highest number of clones in the cluster (see Table 1 for farm labeling). The province (labelled in number; see Table 1) with the highest number of clones in the cluster is highlighted with bold and italics
A comparison of the 11 optimal clusters with the inferred genetic relationships (Fig. 3) revealed some consistent patterns of variation with respect to variety, district and province. For example, the 16 reference varieties were widely spread over the NJ tree or PCoA plot (Fig. 3), while these varieties were grouped in nine of the 11 optimal clusters (Fig. 4B).
Distribution of identified varieties and inferred clusters
Three unique SSR alleles were found to be associated with varieties Rayong 1, Rayong 7 and Sri Racha 1 from the farm clones and three unique combinations of two SSR alleles were obtained to be associated with varieties Kasetsart 50, Huay Bong 80 and Rayong 11. The numbers of the farm clones positively identified as one of the reference varieties is shown in Table 4 and ranged from 3 to 40. Two varieties Kasetsart 50 and Rayong 7 were cultivated across eight provinces, while the other four varieties were unevenly planted (Table 4). Similarly, the optimal 11 clusters of farm clones inferred using STRUCTURE also had an uneven distribution across the provinces (Fig. 4B). For example, the cluster 6 associated with variety Rayong 1 consisted of the farm clones from only four provinces (i.e., Maha Sarakham, Khon Kaen, Kampaeng Phet and Sa Kaeo), while the cluster 8 associated with Kasetsart 50 had the farm clones from all eight provinces (Fig. 4B). Distribution comparison between Table 4 and Fig. 4B seemed to be consistent. For example, the distribution is consistent between Rayong 1 (in Table 4) and cluster 6 associated with Rayong 1 (Fig. 4B) and between Rayong 11 and cluster 4 associated with Rayong 11.
Linking SSR variation to farming variables
Significant associations of estimated pairwise farm genetic distances were found with pairwise farm geographic distances and with differences of farm elevation, annual rainfall, cassava plantation area, cassava production, farm size and farm age (Fig. 5S). Considering the model of isolation by distance (Rousset 1997) helped removing only the marginal association with farm age (from the significance level of p = 0.048–0.058). Clearly, the more distant the farms were, the more genetically differentiated they were. More SSR variation was detected for the farms at the higher elevation. More SSR variation was harbored in the farms with larger cassava plantation and with larger plantation area. However, the SSR variation would decrease for the farms receiving increased annual rainfall and having larger cassava production. All of these associations were generally weak, explaining only 0.6–3.5 % variation in the linear regressions. However, no significant associations were found between on-farm SSR variation and the differences of annual temperature, annual humidity, or the number of years for cassava cultivation (results not shown).
Significant associations were also found between on-farm SSR variation and five categorical farming variables: soil type and the uses of other crops, early cassava varieties, different fertilizers and hormones (Table S3). The assayed clones representing 21 soil types across the 80 farms accounted for 10.5 % SSR variation. The clones from the farms with the Si Songkhram series had the highest diversity content per sample (SeDC = 0.317), while the clones from three farms with the Kabin Buri series had the lowest diversity content per sample (SeDC = 0.293) (see Table S5). The differences in the number of other crops planted and number of early cassava varieties used in a farm explained 3.5 and 2.1 % SSR variations, respectively (Table S3). Interestingly, the farms with records of using more early cassava varieties and more other crops used displayed a trend of increasing SSR variations in the assayed clones, as revealed with the SeDC estimates (Table S4). Also, the farm uses of different fertilizers and hormones were also associated with increased SSR variation in the cassava sample (Table S4) and explained 2.6 and 1.8 % SSR variations, respectively (Table S3). However, allelic counts and tests revealed non-significant allelic differences among the groups of the cassava clones from the farms using variable numbers of fertilizers and hormones (Table S4).
Further Pearson correlation analyses of these farming variables including farm-specific diversity estimates revealed several significant pairwise variable associations (Table S6). As expected, the sample size at a farm was positively correlated with the allelic count and the number of clusters for a farm with sample representation. The farm allelic count was negatively associated with the farm-specific diversity content per sample but positively associated with the number of clusters for a farm with sample representation. These farm diversity variables were not significantly correlated with other farming history and practices. However, several significant correlations were detected among the farming history and practices. The farm age was positively associated with cassava cultivation years, but negatively correlated with the cassava cultivation area and the farm use of different fertilizers and hormones. The farm size was positively correlated with cassava cultivation area and other crops used. The farm use of different other crops were positively associated with the use of more early cassava varieties and large cassava cultivations. Also, the farms using different fertilizers tended to use different hormones. These correlation results suggest that local farming practices have not significantly influenced cassava genetic diversity at the farm level.
Discussion
Our SSR analysis has revealed that the Thai elite cassava genetic diversity was fortuitously conserved by the farmers through farming with different sets of varieties. The assayed farm clones displayed large SSR variation at the farm level (Tables 1 and 2); substantial differentiation among the 80 farms (19.8 %) and across the eight provinces (11.8 %) (Table 2; Figs. 2 and S2); and significant associations between SSR variation and farm agro-ecological factors or some farming practices (Fig. S5; Tables S3 and S4). However, there was no significant genetic differentiation (0.9 %) between the 266 farm clones and 16 reference varieties (Table 2; Fig. 3). A large regional heterogeneity of cassava clones was found and different sets of varieties were planted on 80 farms across eight provinces (Tables 1 and 4; Fig. 4B). These findings advance our understanding of in situ conservation of plant genetic resources and are encouraging for the current efforts in in situ conservation (Veteläinen et al. 2009; Padulosi et al. 2012).
Genetic diversity and structure
The large SSR variation observed in the farm clones is not surprising, as cassava is an outcrossing species with a multi-locus outcrossing rate estimated at 91.5 % (Silva et al. 2003). Our result is consistent with those reported for cassava germplasm from other countries using SSR markers (e.g., Chavarriaga-Aguirre et al. 1998; Fregene et al. 2003; Rocha et al. 2008; Turyagyenda et al. 2012). The prior defined structural inferences revealed seven groups at the farm level, while the Bayesian inference suggested 11 optimal clusters. These inferences gained some support from the individual association analyses (Fig. 3). The 11 optimal clusters also matched with the genetic ancestry of Thai cassava breeding, in which about 10 ancestral lines were known as the major contributors to the Thai breeding germplasm since the 1930s (Ratanawaraha et al. 2001). One of the 11 optimal clusters (Cluster 10; Fig. 4B) was largely unique to the sweet cassava germplasm. These findings were also consistent with those reported from ISSR-based characterization of Thai cassava germplasm (Sakuanrungsirikul et al. 2008).
Evidence for on-farm diversity conservation
There are two lines of evidence which support on-farm conservation of Thai elite cassava germplasm. First, a non-significant SSR differentiation was found between the farm clones and reference varieties (Table 2). Second, the reference varieties were genetically spread widely over the PCoA plot of the farm clones and dispersed broadly into the NJ tree (Fig. 3). However, if only allelic richness is considered, the varieties appeared to display slightly more SSR alleles (365) than expected (356) given the sample size (Table S4). Such an allelic difference is consistent with the estimates of the group-specific entropy-based diversity content per sample (0.3059 and 0.3073 for the farm clones and reference varieties, respectively; Table S4). This small discrepancy may signal that some allelic loss had occurred in the farm clones over time, even though the overall genetic diversity (i.e., allelic richness and evenness) was conserved.
Evidence for regional heterogeneity of cassava clones
The farm use survey confirmed that eight of the 16 reference varieties (R2, R5, R9, R11, R72, K50, H60, H) were cultivated on the farms (Table S1). The direct genetic identification of the farm clones based on unique SSR allele(s) of a given variety confirmed the use on farm of another four varieties (R1, R7, H80, SR) (Table 4). The genetic structure inferences revealed that four clusters of the cassava clones associated with R60, R90, R3 and M were planted on eight or more farms across the eight provinces (Fig. 4B). These results indicated that the 16 reference varieties were still cultivated on the Thai farms in 2011–2012. Also, the farm use survey indicated that Kasetsart 50 and Huay Bong 60 were widely planted (Table S1) probably due to their higher yields and better adaptations to unfavorable conditions (Sarakarn et al. 2001). Fourteen farms grew three or more varieties and 51 farms planted at least two varieties (results not shown). These research outcomes were consistent with the structural inference that the 16 varieties were not uniformly cultivated across the 80 farms in the eight provinces (Table 4; Fig. 4B), although the exact distribution remains to be determined.
The farming with different sets of reference varieties was more evident in Table 1, where there are a variable number of genetic clusters for a farm. The revealed patterns of SSR variation were consistent with our original prediction on varietal distributions across the surveyed provinces (Wangsomnuk et al. 2013). For example, significant SSR differentiations were found between the reference varieties and those farm clones from 41 of the 80 farms (Fig. S2) or from seven of the eight provinces (Table 3). These findings clearly indicate that local farming with different sets of reference varieties did not significantly influence the genetic diversity at the farm level, even in combination with various farming practices (see the correlation results in Table S6), but had un-intentionally generated a large regional heterogeneity of cassava clones in the Thai cassava farming system.
Linking SSR variation to farming variables provided further support for regional heterogeneity of cassava clones. Significant associations were found between SSR variation and farm geographic distance, farm elevation, annual rainfall, cassava plantation area, cassava production, farm size and farm age (Fig. S5). These findings indicate that different farmers tended to plant different varieties. For example, a distant farm tended to cultivate varieties different from a nearby farm. Also significant genetic differentiation was found among the farm clones associated with different farming practices such as farm soil type and the use of other crops, early cassava varieties, fertilizers and hormones (Table S3). These findings indicate that farmers tended to differentiate the use of Thai varieties based on the farm characteristics such as soil type and fertilizer use. The finding that the farm clones associated with different soil types displayed different levels of diversity content per sample (Table S5) further informed to the presence of local differences in farmers’ variety choices to increase cassava production in response to industrial needs.
Limitations of the diversity investigation
Our investigation focused on the analysis of the cassava genetic diversity at different geographic levels (farm, district, and province) and did not examine intra-varietal polymorphism and its local maintenance in detail like those of the village-based investigations (Elias et al. 2001; Sardos et al. 2008; Duputié et al. 2009). It is possible that the assayed clones might have accumulated fixed somatic mutations since the 1970s. We did not examine the germplasm exchanges among farmers and the incorporation of volunteer seedlings from sexual reproduction into the farming system, and the detailed contribution of gene flow and recombination remains unknown. However, it is certain that these genetic factors had contributed to local diversity maintenance, given the findings that few fully identical SSR profiles were found between the farm clones and reference varieties and some inferred clusters (shown either in Fig. 3A, B) did not have a reference variety. We did not associate the detected SSR variations with cultural diversity (e.g., see Delêtre et al. 2011), and it is possible that different cultural groups in the study region contributed to variable farm variety choices. Our SSR analysis for clone abundance and distribution (e.g., Tables 1 and 4) would be more informative if the genetic identity and background of the farm clones were known with certainty and if hybrid clones were identified. Our farm use survey focused on the cassava production in response to industrial demands, but did not include the questions associated with farmers’ criteria for choosing and retaining varieties to assess the difference in farmers’ preferences. Some data collected on the farms and farming practices were not ideal and less accurate and precise. For example, the number of historical cassava varieties planted on a farm was only a rough estimate by the farmer and may not be the true count of previous cultivations. The same is also true for the collection of farm climate and soil data. All of these issues may have diluted the resolution of the association analyses. Our investigation sampled only one data point in time from the cassava farming system and temporal evaluation is needed to determine the long-term conservation effectiveness. We did not address the risk of local genetic homogenization (i.e., disappearance of early introductions or local landraces; see Almekinders et al. 1994) due to the lasting adoption of elite varieties, as information on early introductions is incomplete, and we cannot exclude that the farm clones comprised early introductions.
Implications for in situ conservation
Without an official in situ conservation program for cassava in Thailand, the current cassava farming system has fortuitously conserved the Thai elite cassava genetic diversity. This is most likely the result of farmers maintaining different sets of varieties in different regions of Thailand, in response to local heterogeneities in climate and soil type, as well as industrial demands on cassava. The finding from such an unregulated conservation system is encouraging for the current efforts in in situ conservation of plant genetic resources (Veteläinen et al. 2009; Padulosi et al. 2012). Specifically, a region-focused and farm-based program for conserving the Thai adapted cassava germplasm, along with the ex situ conservation effort, could be developed in Thailand. Specific on-farm conservation strategies could be derived using the genetic structures inferred at the levels of farm, district and province. Farms should be selected based on farm diversity estimates from different provinces to maximize regional diversity. The soil type could also be considered as a good adaptation indicator (Tables S3 and S5). Some farming practices could be used to monitor on-farm diversity, as they were significantly associated with cassava SSR variations (Fig. S5). Such a systematic and long-term program should be more effective than the unregulated one, as the current findings may represent only the outcome for the boom of Thai cassava production over the last 40 years.
What role an unregulated conservation system should play in in situ conservation remains to be explored. We do not know how general the Thai cassava finding is with respect to other tropical crops in other countries. Some important questions are whether such an unregulated system is stable over time and whether it can remain effective under various factors such as changing demands from the industry, climate change, and local needs for subsistence. However, it is certain that unregulated conservation systems could provide a null conservation model for evaluating and monitoring the compared effectiveness of regulated conservation systems. An unique feature of the Thai cassava farming is the self-organization of regional heterogeneity of variety use by local farmers, which is instrumental to in situ conservation designs. More could be learnt from unregulated conservation systems for the better understanding and development of an effective in situ conservation program.
References
Almekinders CJM, Louwaars NP, de Bruijn GH (1994) Local seed systems and their importance for an improved seed supply in developing countries. Euphytica 78:207–216
Altieri MA, Merrick LC (1987) In situ conservation of crop genetic resources through maintenance of traditional farming systems. Econ Bot 41:86–96
Arnaud-Haond S, Duarte CM, Alberto F, Serrao EA (2007) Standardizing methods to address clonality in population studies. Mol Ecol 24:5115–5139
ASEAN Economic Community (2013) Thailand tapioca strategic framework under the AEC. Bangkok, Thailand. http://www.thai-aec.com/71. Accessed 3 April 2014
Awoleye F, van Duren M, Dolezel J, Novak FJ (1994) Nuclear DNA content and in vitro induced somatic polyploidization cassava (Manihot esculenta Crantz) breeding. Euphytica 76:195–202
Brush SB (1991) A farmer-based approach to conserving crop germplasm. Econ Bot 45:153–165
Brush SB (2000) Genes in the field: on-farm conservation of crop diversity. IPGRI, Rome
CBD (2002) Global strategy for plant conservation. Secretariat of the Convention on Biological Diversity, Montreal. http://www.cbd.int/doc/publications/pc-brochure-en.pdf. Accessed 3 April 2014
Chavarriaga-Aguirre P, Maya M, Bonierbale M, Kresovich S, Fregene M, Tohme J, Kochert G (1998) Microsatellites in cassava (Manihot esculenta Crantz): discovery, inheritance and variability. Theor Appl Genet 97:493–501
Cock JH (1985) Cassava: new potential for a neglected crop. Westview Press, Boulder
Debouck D, Dominique D, Alexandra J, Hershey C, Llerme R (2011) Conservation of cassava genetic resource. http://cropgenebank.sgrp.cgiar.org/index.php?option=com_content&view=article&id=113&Itemid=213. Accessed 3 April 2014
Delêtre M, McKey DB, Hodkinson TR (2011) Marriage exchanges, seed exchanges and the dynamics of crop diversity. Proc Natl Acad Sci USA 108:18249–18254
Department of Agriculture (2013) Bangkok, Thailand. http://210.246.186.198/~cassava/var/var.html. Accessed 3 April 2014
Duputié A, Massol F, David P, Haxaire C, McKey D (2009) Traditional Amerindian cultivators combine directional and ideotypic selection for sustainable management of cassava genetic diversity. J Evol Biol 22:1317–1325
Eklund A (2011) Squash: color-based plots for multivariate visualization. R Package version 1.0.1. http://www.cbs.dtu.dk/~eklund/squash/
Elias M, Panaud O, Robert T (2000) Assessment of genetic variability in a traditional cassava (Manihot esculenta Crantz) farming system, using AFLP markers. Heredity 85:219–230
Elias M, Penet L, Vindry P, Mckey D, Panaud O, Robert T (2001) Unmanaged sexual reproduction and the dynamics of genetic diversity of a vegetatively propagated crop plant, cassava (Manihot esculenta Crantz), in a traditional farming system. Mol Ecol 10:1895–1907
Elias M, Mühlen GS, McKey D, Roa AC, Tohme J (2004) Genetic diversity of traditional South American landraces of cassava (Manihot esculenta Crantz): an analysis using microsatellites. Econ Bot 58:242–256
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
FAO (2010) The second report on the state of the world’s plant genetic resources for food and agriculture. Italy, Rome
FAO/IFAD (2001) The global cassava development strategy and implementation plan. Proceedings of the validation forum on the global cassava development strategy. Rome, 26–28 April 2000
Fregene M, Suarez M, Mkumbira J, Kulembeka H, Ndedya E, Kulaya A, Mitchel S, Gullberg U, Rosling H, Dixon A, Dean R, Kresovich S (2003) Simple sequence repeat marker diversity in cassava landraces: genetic diversity and differentiation in an asexually propagated crop. Theor Appl Genet 107:1083–1093
Fu YB (2010) FPTest: a SAS routine for testing allelic counts. Mol Ecol Resour 10:389–392
Hershey CH, Debouck D (2010) A global conservation strategy for cassava and wild Manihot species. A summary of stakeholder deliberations and recommendations prepared for the Global Crop Diversity Trust. http://www.croptrust.org/documents/cropstrategies/cassava%20strategy.pdf. Accessed 3 April 2014
Jarvis DI, Padoch C, Cooper HD (2007) Managing biodiversity in agricultural ecosystems. Columbia University Press, New York, pp 1–492
Jarvis DI, Brown AHD, Cuong PH, Panduro LC, Moreno LL, Gyawali S, Tanto T, Sawadogo M, Mar I, Sadiki M, Hue NTN, Reyes LA, Balma D, Bajracharya J, Castillo F, Rijal D, Belqadi L, Rana R, Saidi S, Ouedraogo J, Zangre R, Rhrib K, Chavez JL, Schoen D, Sthapit B, Santis PD, Fadda C, Hodgkin T (2008) A global perspective of the richness and evenness of traditional crop-variety diversity maintained by farming communities. Proc Natl Acad Sci USA 105:5326–5331
Jennings DL (1995) Manihot esculenta (Euphorbiaceae). In: Smartt J, Simmonds NW (eds) Evolution of crop plants. Wiley, New York, pp 128–132
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Kunkeaw S, Yoocha T, Sraphet S, Boonchanawiwat A, Boonseng O, Lightfoot DA, Triwitayakorn K, Tangphatsornruang S (2010) Construction of a genetic linkage map using simple sequence repeat markers from expressed sequence tags for cassava (Manihot esculenta Crantz). Mol Breed 27:67–75
Maxted N, Ford-Lloyd BV, Hawkes JG (1997) Complementary conservation strategies. In: Maxted N, Ford-Lloyd BV, Hawkes JG (eds) Plant genetic conservation: the in situ approach. Chapman & Hall, London, pp 20–55
Mba REC, Stephenson P, Edwards K, Melzer S, Nkumbira J, Gullberg U, Apel K, Gale M, Tohme J, Fregene M (2001) Simple sequence repeat (SSR) marker survey of the cassava (Manihot esculenta Crantz) genome: towards an SSR-based molecular genetic map of cassava. Theor Appl Genet 102:21–31
McKey D, Elias M, Pujol B, Duputie A (2010) The evolutionary ecology of clonally propagated domesticated plants. New Phytol 186:318–332
Montero-Rojas M, Correa AM, Siritunga D (2011) Molecular differentiation and diversity of cassava (Manihot esculenta) taken from 162 locations across Puerto Rico and assessed with microsatellite markers. AoB Plants 2011 plr010
Nevo E, Fu YB, Pavlicek T, Khalifa S, Tavasi M, Beiles A (2012) Evolution of wild cereals during 28 years of global warming in Israel. Proc Natl Acad Sci USA 109:3412–3415
Office of Agricultural Economics (2013) Cassava production statistics. Bangkok, Thailand. http://www.oae.go.th/download/prcai/DryCrop/cassava53-55.pdf. Accessed 3 April 2014
Office of Soil Resources Survey and Research (2011a) Soil map of Thailand. Land Development Department, Ministry of Agriculture and Cooperatives. Bangkok, Thailand. http://oss101.ldd.go.th/web_th_soilseries/INDEX_th_series.htm. Accessed 3 April 2014
Office of Soil Resources Survey and Research (2011b) Soil of Thailand. Land Development Department, Ministry of Agriculture and Cooperatives. Bangkok, Thailand. http://oss101.ldd.go.th/thaisoils_museum/INDEX.HTM. Accessed 3 April 2014
Office of Soil Survey and Land Use Planning (2004a) Characterization of established soil series in the Central Plain region of Thailand reclassified according to Soil Taxonomy 2003. Land Development Department, Ministry of Agriculture and Cooperatives, Bangkok
Office of Soil Survey and Land Use Planning (2004b) Characterization of established soil series in the North and Central Highland region of Thailand reclassified according to Soil Taxonomy 2003. Land Development Department, Ministry of Agriculture and Cooperatives, Bangkok
Office of Soil Survey and Land Use Planning (2004c) Characterization of established soil series in the Northeast region of Thailand reclassified according to Soil Taxonomy 2003. Land Development Department, Ministry of Agriculture and Cooperatives, Bangkok
Padulosi S, Bergamini N, Lawrence T (2012) On farm conservation of neglected and underutilized species: status, trends and novel approaches to cope with climate change: proceedings of an International Conference, Frankfurt; 14–16 June, 2011; Biodiversity International, Rome, pp 1–293
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pritchard J, Stephens M, Donnelly P (2000) Influence of population structure using multilocus genotype data. Genetics 155:945–959
R Development Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. ISBN 3-900051-07-0. http://www.R-project.org. Accessed 3 April 2014
Raji AA, Anderson JV, Kolade OA, Ugwu CD, Dixon AG, Ingelbrecht IL (2009) Gene-based microsatellites for cassava (Manihot esculenta Crantz): prevalence, polymorphisms, and cross-taxa utility. BMC Plant Biol 9:118
Ratanawaraha C, Senanarong N, Suriyapan P (2001) Status of cassava in Thailand: implications for future research and development. In: A review of cassava in Asia with country case studies on Thailand and Viet Nam. Proceedings of the validation forum on the global cassava development strategy. Rome; 26–28 April 2000. http://www.fao.org/docrep/009/y1177e/Y1177E04.htm. Accessed 3 April 2014
Reyes-Valdes MH, Williams CG (2005) An entropy-based measure of founder informativeness. Genet Res 85:81–88
Rocha O, Zaldimar M, Castro L, Castro E, Barrantes R (2008) Microsatellite variation of cassava (Manihot esculenta Crantz) in home gardens of Chibchan Amerindians from Costa Rica. Conserv Genet 9:107–118
Rohlf FJ (1997) NTSYS-pc 2.1. Numerical taxonomy and multivariate analysis system. Exeter Software, Setauket, NY
Rojanaridpiched C (1988) Cassava varietal improvement at Kasetsart University, Thailand. In: Howeler RH and Kawano K (eds) Cassava breeding and agronomy research in Asia: proceedings of a regional workshop; Oct 26–28, 1987; Rayong, Thailand, pp 21–26
Rojanaridpiched C, Vichukit V, Sarobol E, Changlek P (2007) Breeding and dissemination of new cassava varieties in Thailand. In: Howeler RH (ed) Cassava research and development in Asia: exploring new opportunities for an ancient Crop. In: Proceedings of the 7th regional cassava workshop; Oct. 28–Nov. 1, 2002; Bangkok, Thailand
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Sakuanrungsirikul S, Sarawat P, Wongtiem P, Sarakarn S, Khonsantear V, Khantahad R (2008) Molecular characterization of the CIAT cassava core collection and the Thai and Lao germplasm collection using ISSR-Touchdown PCR. In: Howeler RH (ed) A new future for cassava in Asia: its use as food, feed and fuel to benefit the poor. Proceedings of the 8th regional workshop; Oct 20–24, 2008; Vientiane, Lao PDR, pp 184–189
Salick J, Celinesse N, Knapp S (1997) Indigenous diversity of cassava: generation, maintenance, use and loss among the Amuesha, Peruvian Upper Amazon. Econ Bot 51:6–19
Sambatti J, Martins P, Ando A (2001) Folk taxonomy and evolutionary dynamics of cassava: a case study in Ubatuba, Brazil. Econ Bot 55:93–105
Sarakarn S, Limsila A, Watananonta W, Suparharn D, Suriyapan P (2001) Cassava breeding and varietal dissemination in Thailand-Major achievements during the past 25 years. In: Howeler RH and Tan SL (eds) Cassava’s potential in Asia in the 21st century: present situation and future research and development needs. Proceedings of the 6th regional workshop; Feb 21–25, 2000; Ho Chi Minh City, Vietnam, pp 161–166
Sarakarn S, Limsila A, Suparhan D, Wongtiem P, Hansetasuk J, Watananonta W (2007) Cassava germplasm conservation and crop improvement in Thailand. In: Howeler RH (ed) Cassava research and development in Asia: exploring new opportunities for an ancient crop. Proceedings of the 7th regional cassava workshop; Oct. 28–Nov. 1, 2002; Bangkok, Thailand, pp 58–66
Sardos J, Mackey E, Duval MF, Malapa R, Noyer JL, Lebot V (2008) Evolution of cassava (Manihot esculenta Crantz) after recent introduction into a South Pacific Island system: the contribution of sex to the diversification of a clonally propagated crop. Genome 51:912–921
SAS Institute Inc (2008) The SAS system for windows V9.2. SAS Institute Incorporated, Cary, NC
Silva R, Bandel G, Martins P (2003) Mating system in an experimental garden composed of cassava (Manihot esculenta Crantz) ethnovarieties. Euphytica 134:127–135
Siqueira MVBM, Queiroz-Silva JR, Bressan EA, Borges A, Pereira KJC, Pinto JG, Veasey EA (2009) Genetic characterization of cassava (Manihot esculenta) landraces in Brazil assessed with simple sequence repeats. Genet Mol Biol 32:104–110
Swofford DL (1998) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts
Thai Meteorological Department (2012) Bangkok, Thailand. http://www.tmd.go.th/. Assessed on 3 April 2014
Thomas M, Demeulenaere E, Dawson JC, Khan AR, Galic N, Jouanne-Pin S, Remoue C, Bonneuil C, Goldringer I (2012) On-farm dynamic management of genetic diversity: the impact of seed diffusions and seed saving practices on a population-variety of bread wheat. Evol Appl 5:779–795
Turyagyenda LF, Kizito EB, Ferguson ME, Baguma Y, Harvey JW, Gibson P, Wanjala BW, Osiru DSO (2012) Genetic diversity among farmer-preferred cassava landraces in Uganda. Afr Crop Sci J 20(Suppl 1):15–30
Veteläinen M, Negri V, Maxted N (2009) European landraces: on farm conservation, management and use. Biodiversity Technical Bulletin No. 15. Bioversity International, Rome, Italy, pp 1–344
Wangsomnuk P, Ruttawat B, Wongtiem P (2013) Identification of genetically distinct cassava clones from on-farm plantations to widen the Thai cassava breeding gene pool. Am J Plant Sci 4:1574–1583
Acknowledgments
The authors thank all farmers who participated in the farm survey and cassava sampling; Sitteera Chawnue for her assistance on farm clone collection; Tossapol Pholcharee and Patchanee Arpornrat for helping with the farm survey data; Dr. Prapit Wongtiem, Dr. Pinich Wangsomnuk and Sutti Thawanthaisong for their technical advice; all village leaders and the related staffs of the Provincial Department of Agriculture for their kind cooperation for this study; Thai Meteorological Department and Land Development Department for providing the secondary data; Rayong Field Crop Research Center, Khon Kaen Field Crops Research Center, and Department of Agriculture for providing the variety clones; and two journal reviewers for their helpful comments on the early version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Fu, YB., Wangsomnuk, P.P. & Ruttawat, B. Thai elite cassava genetic diversity was fortuitously conserved through farming with different sets of varieties. Conserv Genet 15, 1463–1478 (2014). https://doi.org/10.1007/s10592-014-0631-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-014-0631-y