Abstract
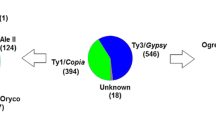
Capsicum annuum is a species that has undergone an expansion of the size of its genome caused mainly by the amplification of repetitive DNA sequences, including mobile genetic elements. Based on information obtained from sequencing the genome of pepper, the estimated fraction of retroelements is approximately 81%, and previous results revealed an important contribution of lineages derived from Gypsy superfamily. However, the dynamics of the retroelements in the C. annuum genome is poorly understood. In this way, the present work seeks to investigate the phylogenetic diversity and genomic abundance of the families of autonomous (complete and intact) LTR retroelements from C. annuum and inspect their distribution along its chromosomes. In total, we identified 1151 structurally full-length retroelements (340 Copia; 811 Gypsy) grouped in 124 phylogenetic families in the base of their retrotranscriptase. All the evolutive lineages of LTR retroelements identified in plants were present in pepper; however, three of them comprise 83% of the entire LTR retroelements population, the lineages Athila, Del/Tekay, and Ale/Retrofit. From them, only three families represent 70.8% of the total number of the identified retroelements. A massive family-specific wave of amplification of two of them occurred in the last 0.5 Mya (GypsyCa_16; CopiaCa_01), whereas the third is more ancient and occurred 3.0 Mya (GypsyCa_13). Fluorescent in situ hybridization performed with family and lineage-specific probes revealed contrasting patterns of chromosomal affinity. Our results provide a database of the populations LTR retroelements specific to C. annuum genome. The most abundant families were analyzed according to chromosome insertional preferences, suppling useful tools to the design of retroelement-based markers specific to the species.






Similar content being viewed by others
Abbreviations
- C:
-
centromeric chromosome portion
- CTAB:
-
cetyl trimethyl ammonium bromide
- FISH:
-
fluorescent in situ hybridization
- IP:
-
interstitial-proximal chromosome portion
- IT:
-
interstitial-terminal chromosome portion
- LTR:
-
long terminal repeats
- Mya:
-
millions of years ago
- P:
-
proximal chromosome portion
- PCR:
-
polymerase chain reaction
- RH:
-
RNase-H
- RT:
-
retrotranscriptase
- T:
-
terminal chromosome portion
References
Anderson, SN, Stitzer, MC, Brohammer, AB, Zhou, P, Noshay, JM, O’Connor, CH, … Springer, NM (2019) Transposable elements contribute to dynamic genome content in maize. Plant J, 100(5), 1052–1065. https://doi.org/10.1111/tpj.14489
Argout X, Salse J, Aury JM, Guiltinan MJ, Droc G, Gouzy J et al (2011) The genome of Theobroma cacao. Nat Genet 43(2):101–108. https://doi.org/10.1038/ng.736
Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, … Bennetzen JL (2009) Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genetics 5(11). https://doi.org/10.1371/journal.pgen.1000732
Beliveau, BJ, Joyce, EF, Apostolopoulos, N, Yilmaz, F, Fonseka, CY, McCole, RB, ... & Wu, CT (2012) Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci, 109(52), 21301–21306. https://doi.org/10.1073/pnas.1213818110
Belyayev A (2014) Bursts of transposable elements as an evolutionary driving force. J Evol Biol 27(12):2573–2584. https://doi.org/10.1111/jeb.12513
Benachenhou F, Sperber GO, Bongcam-Rudloff E, Andersson G, Boeke JD, Blomberg J (2013) Conserved structure and inferred evolutionary history of long terminal repeats (LTRs). Mob DNA 4(1):1–16. https://doi.org/10.1186/1759-8753-4-5
Bennetzen JL (1996) The Mutator transposable element family of maize. Genet Eng 13:1–37. https://doi.org/10.1016/0966-842X(96)10042-1
Bennetzen JL (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42(1):251–269. https://doi.org/10.1023/A:1006344508454
Bennetzen JL (2002) Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica 115(1):29–36. https://doi.org/10.1023/A:1016015913350
Bennetzen JL, Wang H (2014) The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol 65(1):505–530. https://doi.org/10.1146/annurev-arplant-050213-035811
Beulé T, Agbessi MDT, Dussert S, Jaligot E, Guyot R (2015) Genome-wide analysis of LTR-retrotransposons in oil palm. BMC Genomics 16(1):795. https://doi.org/10.1186/s12864-015-2023-1
Biémont C, Vieira C (2006) Genetics: junk DNA as an evolutionary force. Nature 443(7111):521–524. https://doi.org/10.1038/443521a
Bolger AM, Lohse M, Usadel B (2014) Genome analysis Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bourgeois Y, Boissinot S (2019) On the population dynamics of junk: a review on the population genomics of transposable elements. Genes. 10:419. https://doi.org/10.3390/genes10060419
Bowen NJ, McDonald JF (2001) Drosophila euchromatic LTR retrotransposons are much younger than the host species in which they reside. Genome Res 11(9):1527–1540. https://doi.org/10.1101/gr.164201
Boyle S, Rodesch MJ, Halvensleben HA, Jeddeloh JA, Bickmore WA (2011) Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosom Res 19(7):901–909. https://doi.org/10.1007/s10577-011-9245-0
Braz GT, He L, Zhao H, Zhang T, Semrau K, Rouillard JM et al (2018) Comparative oligo-FISH mapping: an efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 208(2):513–523. https://doi.org/10.1534/genetics.117.300344
Cavrak VV, Lettner N, Jamge S, Kosarewicz A, Bayer LM, Scheid OM (2014) How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet 10(1):e1004115. https://doi.org/10.1371/journal.pgen.1004115
Datson PM, Murray BG (2006) Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism? Chromosome. https://doi.org/10.1007/s10577-006-1092-z
De Assis R, Baba VY, Cintra LA, Goncalves LSA, Rodrigues R, Vanzela ALL (2020) Genome relationships and ltr-retrotransposon diversity in three cultivated Capsicum L. (Solanaceae) species. BMC Genomics 21(1):237. https://doi.org/10.1186/s12864-020-6618-9
De Jong JH, Fransz P, Zabel P (1999) High resolution FISH in plants–techniques and applications. Trends Plant Sci 4(7):258–263. https://doi.org/10.1016/S1360-1385(99)01436-3
De Souza TB, Chaluvadi SR, Johnen L, Marques A, González-Elizondo MS, Bennetzen JL, Vanzela ALL (2018) Analysis of retrotransposon abundance, diversity and distribution in holocentric Eleocharis (Cyperaceae) genomes. Ann Bot 122(2):279–290. https://doi.org/10.1093/aob/mcy066
Devos KM, Brown JK, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res 12(7):1075–1079. https://doi.org/10.1101/gr.132102
Domingues DS, Cruz GMQ, Metcalfe CJ, Nogueira FTS, Vicentini R, …, Van Sluys, MA (2012) Analysis of plant LTR-retrotransposons at the fine-scale family level reveals individual molecular patterns. BMC Genomics, 13(1), 137. https://doi.org/10.1186/1471-2164-13-137
Du, J, Tian, Z, Hans, CS, Laten, HM, Cannon, SB, Jackson, SA., … Ma, J (2010) Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J, 63(4), 584–598. https://doi.org/10.1111/j.1365-313X.2010.04263.x
Dubcovsky J, Dvorák J (1995) Ribosomal RNA multigene loci: nomads of the Triticeae genomes. Genetics 140(4):367–377
Esposito S, Barteri F, Casacuberta J, Mirouze M, Carputo D, Aversano R (2019) LTR-TEs abundance, timing and mobility in Solanum commersonii and S. tuberosum genomes following cold-stress conditions. Planta 1:1781–1787. https://doi.org/10.1007/s00425-019-03283-3
Feschotte C, Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41(1):331–368. https://doi.org/10.1146/annurev.genet.40.110405.090448
Gaiero P, Vaio M, Peters SA, Eric Schranz M, de Jong H, Speranza PR (2019) Comparative analysis of repetitive sequences among species from the potato and the tomato clades. Ann Bot 123:521–532. https://doi.org/10.1093/aob/mcy186
Galindo-González L, Mhiri C, Deyholos MK, Grandbastien MA (2017) LTR-retrotransposons in plants: engines of evolution. Gene 626:14–25. https://doi.org/10.1016/j.gene.2017.04.051
Gao X, Hou Y, Ebina H, Levin HL, Voytas DF (2008) Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res 18(3):359–369. https://doi.org/10.1101/gr.7146408
Gouy M, Guindon S, Gascuel O (2010) SeaView Version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27(2):221–224. https://doi.org/10.1093/molbev/msp259
Grandbastien MA (1998) Activation of plant retrotransposons under stress conditions. Trends Plant Sci 3(5):181–187. https://doi.org/10.1016/S1360-1385(98)01232-1
Gribbon, BM, Pearce, SR, Kalendar, R, Schulman, AH, Paulin, L, Jack, P, ... & Flavell, AJ (1999) Phylogeny and transpositional activity of Ty1-copia group retrotransposons in cereal genomes. Mol Gen Genet MGG, 261(6), 883–891. https://doi.org/10.1007/PL00008635
Havecker ER, Gao X, Voytas DF (2004) The diversity of LTR retrotransposons. Genome Biol BioMed Central. https://doi.org/10.1186/gb-2004-5-6-225
Hirochika H, Hirochika R (1993) Tyl-copia group retrotransposons as ubiquitous components of plant genomes. Jpn J Genet. https://doi.org/10.1266/jjg.68.35
Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M (1996) Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci 93(15):7783–7788. https://doi.org/10.1073/pnas.93.15.7783
Huang Y, Luo L, Hu X, Yu F, Yang Y, Deng Z et al (2017) Characterization, genomic organization, abundance, and chromosomal distribution of Ty1-copia retrotransposons in Erianthus arundinaceus. Front Plant Sci 8:924. https://doi.org/10.3389/fpls.2017.00924
Jarillo, J. A., Pineiro, M., Cubas, P., Martinez-Zapater, J. M., Jarillo, J. A., Pineiro, M., … Martinez-Zapater, J. M. (2009). Chromatin remodeling in plant development. Int J Dev Biol 53(8-9–10), 1581–1596. https://doi.org/10.1387/ijdb.072460jj
Jääskeläinen M, Chang W, Moisy C, Schulman AH (2013) Retrotransposon BARE displays strong tissue-specific differences in expression. New Phytol 200(4):1000–1008. https://doi.org/10.1111/nph.12470
Jaillon, O, Aury, JM, Noel, B, Policriti, A, Clepet, C, Casagrande, A, … Wincker, P (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature, 449(7161), 463–467. https://doi.org/10.1038/nature06148
Jiang, N, Bao, Z, Temnykh, S, Cheng, Z, Jiang, J, Wing, RA, ... & Wessler, SR (2002a) Dasheng: a recently amplified nonautonomous long terminal repeat element that is a major component of pericentromeric regions in rice. Genetics, 161(3), 1293–1305. https://doi.org/10.1093/genetics/161.3.1293
Jiang N, Jordan IK, Wessler SR (2002b) Dasheng and RIRE2. A nonautonomous long terminal repeat element and its putative autonomous partner in the rice genome. Plant Physiol 130(4):1697–1705. https://doi.org/10.1104/pp.015412
Jiang SY, Ramachandran S (2013) Genome-wide survey and comparative analysis of LTR retrotransposons and their captured genes in rice and sorghum. PLoS One 8(7):e71118. https://doi.org/10.1371/journal.pone.0071118
Kalendar R, Vicient CM, Peleg O, Anamthawat-Jonsson K, Bolshoy A, Schulman AH (2004) Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 166(3):1437–1450. https://doi.org/10.1534/genetics.166.3.1437
Kato A, Lamb JC, Birchler JA (2004) Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci U S A 101:13554–13559. https://doi.org/10.1073/pnas.0403659101
Kejnovsky E, Kubat Z, Macas J, Hobza R, Mracek J, Vyskot B (2006) Retand: a novel family of gypsy-like retrotransposons harboring an amplified tandem repeat. Mol Gen Genomics 276(3):254–263. https://doi.org/10.1007/s00438-006-0140-x
Kijima TE, Innan H (2009) On the estimation of the insertion time of LTR retrotransposable elements. Mol Biol Evol 27(4):896–904. https://doi.org/10.1093/molbev/msp295
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33(1):479–532. https://doi.org/10.1146/annurev.genet.33.1.479
Kriedt RA, Cruz GMQ, Bonatto SL, Freitas LB (2014) Novel transposable elements in Solanaceae: evolutionary relationships among Tnt1-related sequences in wild Petunia species. Plant Mol Biol Report 32(1):142–152. https://doi.org/10.1007/s11105-013-0626-8
Levan A, Fredga K, Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52(2):201–220. https://doi.org/10.1111/j.1601-5223.1964.tb01953.x
Li, SF, Guo, YJ, Li, JR, Zhang, DX, Wang, B X, Li, N, …, Gao, WJ (2019) The landscape of transposable elements and satellite DNAs in the genome of a dioecious plant spinach (Spinacia oleracea L.). Mobile DNA, 10(1), 3. https://doi.org/10.1186/s13100-019-0147-6
Llorens C, Muñoz-Pomer A, Bernad L, Botella H, Moya A (2009) Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol Direct 4:41. https://doi.org/10.1186/1745-6150-4-41
Llorens, C, Futami, R, Covelli, L, Domínguez-Escribá, L, Viu, JM, Tamarit, D, … Moya, A (2011) The Gypsy Database (GyDB) of Mobile Genetic Elements: Release 2.0. Nucleic Acids Res, 39(SUPPL. 1), 38–46. https://doi.org/10.1093/nar/gkq1061
Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14(5):860–869. https://doi.org/10.1101/gr.1466204
Manetti ME, Rossi M, Nakabashi M, Grandbastien MA, Van Sluys MA (2009) The Tnt1 family member Retrosol copy number and structure disclose retrotransposon diversification in different Solanum species. Mol Gen Genomics 281(3):261–271. https://doi.org/10.1007/s00438-008-0408-4
Marcon HS, Domingues DS, Silva JC, Borges RJ, Matioli FF, de Mattos Fontes MR, Marino CL (2015) Transcriptionally active LTR retrotransposons in Eucalyptus genus are differentially expressed and insertionally polymorphic. BMC Plant Biol 15(1):1–16. https://doi.org/10.1186/s12870-015-0550-1
Maupetit-Mehouas S, Vaury C (2020) Transposon reactivation in the germline may be useful for both transposons and their host genomes. Cells. 9(5):1172. https://doi.org/10.3390/cells9051172
Melayah, D, Lim, KY, Bonnivard, E, Chalhoub, B, Dorlhac De Borne, F, Mhiri, C, … Grandbastien, MA (2004) Distribution of the Tnt1 retrotransposon family in the amphidiploid tobacco (Nicotiana tabacum) and its wild Nicotiana relatives. Biol J Linn Soc, 82(4), 639–649. https://doi.org/10.1111/j.1095-8312.2004.00348.x
Myers T (2001) Rice genome consortium will finish ahead of schedule. Nature 409:752. https://doi.org/10.1038/35057489
Naito, K, Zhang, F, Tsukiyama, T, Saito, H, Hancock, CN, Richardson, AO, … Wessler, SR (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature, 461(7267), 1130–1134. https://doi.org/10.1038/nature08479
Nellåker, C, Keane, T M, Yalcin, B, Wong, K, Agam, A, Belgard, TG, … Ponting, CP (2012) The genomic landscape shaped by selection on transposable elements across 18 mouse strains. Genome Biol, 13(6). https://doi.org/10.1186/gb-2012-13-6-r45
Neumann, P, Navrátilová, A, Koblížková, A, Kejnovsk, E, Hřibová, E, Hobza, R, … MacAs, J (2011) Plant centromeric retrotransposons: a structural and cytogenetic perspective. Mob DNA, 2(1), 1–16. https://doi.org/10.1186/1759-8753-2-4
Neumann P, Novák P, Hoštáková N, Macas J (2019) Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob DNA 10(1):1–17. https://doi.org/10.1186/s13100-018-0144-1
Okonechnikov K, Golosova O, Fursov M, Team U (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28(8):1166–1167. https://doi.org/10.1093/bioinformatics/bts091
Park, M, Jo, SH, Kwon, JK, Park, J, Ahn, JH, Kim, S, … Choi, D (2011) Comparative analysis of pepper and tomato reveals euchromatin expansion of pepper genome caused by differential accumulation of Ty3/Gypsy-like elements. BMC Genomics, 12(1), 85. https://doi.org/10.1186/1471-2164-12-85
Park, M, Park, J, Kim, S, Kwon, JK, Park, HM, Bae, IH, … Choi, D (2012) Evolution of the large genome in Capsicum annuum occurred through accumulation of single-type long terminal repeat retrotransposons and their derivatives. Plant J, 69(6), 1018–1029. https://doi.org/10.1111/j.1365-313X.2011.04851.x
Paz RC, Rendina González AP, Ferrer MS, Masuelli RW (2015) Short-term hybridisation activates Tnt1 and Tto1 Copia retrotransposons in wild tuber-bearing Solanum species. Plant Biol 17(4):860–869. https://doi.org/10.1111/plb.12301
Paz RC, Kozaczek ME, Rosli HG, Andino NP, Sanchez-Puerta MV (2017) Diversity, distribution and dynamics of full-length Copia and Gypsy LTR retroelements in Solanum lycopersicum. Genetica 145(4–5):417–430. https://doi.org/10.1007/s10709-017-9977-7
Pearce SR, Pich U, Harrison G, Flavell AJ, Heslop-Harrison JS, Schubert I, Kumar A (1996) The Ty1-copia group retrotransposons of Allium cepa are distributed throughout the chromosomes but are enriched in the terminal heterochromatin. Chromosom Res 4(5):357–364. https://doi.org/10.1007/BF02257271
Petit, M, Lim, KY, Julio, E, Poncet, C, De Borne, FD, Kovarik, A, ... & Mhiri, C (2007) Differential impact of retrotransposon populations on the genome of allotetraploid tobacco (Nicotiana tabacum). Mol Gen Genomics, 278(1), 1–15. https://doi.org/10.1007/s00438-007-0226-0
Piegu, B., Guyot, R., Picault, N., Roulin, A., Sanyal, A., Kim, H., … Panaud, O. (2006). Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res, 21(7), 1201. https://doi.org/10.1101/gr.5290206.of
Qin, C, Yu, C, Shen, Y, Fang, X, Chen, L, Min, J & Cheng, J (2014) Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc Natl Acad Sci 111(14). https://doi.org/10.1073/pnas.1400975111
Qiu F, Ungerer MC (2018) Genomic abundance and transcriptional activity of diverse gypsy and copia long terminal repeat retrotransposons in three wild sunflower species. BMC Plant Biol 18(1):1–8. https://doi.org/10.1186/s12870-017-1223-z
Raskina O, Belyayev A, Nevo E (2004) Quantum speciation in Aegilops: molecular cytogenetic evidence from rDNA cluster variability in natural populations. Proc Natl Acad Sci U S A 101(41):14818–14823. https://doi.org/10.1073/pnas.0405817101
Raskina O, Barber JC, Nevo E, Belyayev A (2008) Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogen Genome Res 120(3–4):351–357. https://doi.org/10.1159/000121084
Roa F, Guerra M (2012) Distribution of 45S rDNA sites in chromosomes of plants: structural and evolutionary implications. BMC Evol Biol 12(1):1. https://doi.org/10.1186/1471-2148-12-225
Sabot F, Schulman AH (2006) Parasitism and the retrotransposon life cycle in plants: a hitchhiker’s guide to the genome. Heredity 97(6):381–388. https://doi.org/10.1038/sj.hdy.6800903
SanMiguel, P, Tikhonov, A, Jin, YK, Motchoulskaia, N, Zakharov, D, Melake-Berhan, A, ... & Bennetzen, JL (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science, 274(5288), 765–768
SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nat Genet 20(1):43–45. https://doi.org/10.1038/1695
Schnable, PS, Pasternak, S, Liang, C, Zhang, J, Fulton, L, Graves, TA, … Kumari, S (2009) The B73 maize genome: complexity, diversity, and dynamics. Science, 326(June), 1112–1115. https://doi.org/10.1126/science.1178534
Schulman AH, Kalendar R (2005) A movable feast: diverse retrotransposons and their contribution to barley genome dynamics. Cytogen Genome Res 110(1–4):598–605. https://doi.org/10.1159/000084993
Schwarzacher T, Heslop-Harrison P (2000) Practical in situ hybridization. BIOS Scientific Publishers, Oxford. https://doi.org/10.1023/A:1026756103545
Sharma A, Schneider KL, Presting GG (2008) Sustained retrotransposition is mediated by nucleotide deletions and interelement recombinations. Proc Natl Acad Sci U S A 105(40):15470–15474. https://doi.org/10.1073/pnas.0805694105
Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P (2000) A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res 10(7):908–915. https://doi.org/10.1101/gr.10.7.908
Tam SM, Mhiri C, Vogelaar A, Kerkveld M, Pearce SR, Grandbastien MA (2005) Comparative analyses of genetic diversities within tomato and pepper collections detected by retrotransposon-based SSAP, AFLP and SSR. Theor Appl Genet 110(5):819–831. https://doi.org/10.1007/s00122-004-1837-z
Tam SM, Lefebvre V, Palloix A, Sage-Palloix AM, Mhiri C, Grandbastien MA (2009) LTR-retrotransposons Tnt1 and T135 markers reveal genetic diversity and evolutionary relationships of domesticated peppers. Theor Appl Genet 119(6):973–989. https://doi.org/10.1007/s00122-009-1102-6
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599. https://doi.org/10.1093/molbev/msm092
Tomato Genome Consortium, T, DNA Research Institute, K, Sciences, L, company, R., Express Inc, A., Academy of Agriculture, B, …, SrL, Y (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Tomato Genome Consortium https://doi.org/10.1038/nature11119
Valárik M, Bartoš J, Kovářová P, Kubaláková M, De Jong JH, Doležel J (2004) High-resolution FISH on super-stretched flow-sorted plant chromosomes. Plant J 37(6):940–950. https://doi.org/10.1111/j.1365-313X.2003.02010.x
Van de Rijke FM, Florijn RJ, Tanke HJ, Raap K (2000) DNA fiber-FISH staining mechanism. J Histochem Cytochem 48(6):743–745. https://doi.org/10.1177/002215540004800602
Vicient CM, Suoniemi A, Anamthawat-Jonsson K, Tanskanen J, Beharav A, Nevo E, Schulman AH (1999) Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11(9):1769–1784. https://doi.org/10.1105/tpc.11.9.1769
Vicient CM, Jääskeläinen MJ, Kalendar R, Schulman AH (2001) Active retrotransposons are a common feature of grass genomes. Plant Physiol 125(3):1283–1292. https://doi.org/10.1104/pp.125.3.1283
Vicient CM (2010) Transcriptional activity of transposable elements in maize. BMC Genomics 11(1). https://doi.org/10.1186/1471-2164-11-601
Vitte C, Panaud O, Quesneville H (2007) LTR retrotransposons in rice (Oryza sativa, L.): recent burst amplifications followed by rapid DNA loss. BMC Genomics 8:17–21. https://doi.org/10.1186/1471-2164-8-218
Wang, W, Zheng, H, Fan, C, Li, J, Shi, J, Cai, Z, … Wang, J (2006) High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell, 18(8), 1791–1802. https://doi.org/10.1105/tpc.106.041905
Weber, B, Heitkam, T, Holtgräwe, D, Weisshaar, B, Minoche, AE, Dohm, JC, … Schmidt, T (2013) Highly diverse chromoviruses of Beta vulgaris are classified by chromodomains and chromosomal integration. Mob DNA, 4(1), 1–16. https://doi.org/10.1186/1759-8753-4-8
Wicker, T & Keller, B (2007) Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient.pdf, 1072–1081. https://doi.org/10.1101/gr.6214107
Weising K, Nybom H, Wolff K, Kahl G (2005) DNA fingerprinting in plants, 2nd end: principles, methods, and applications. Ann Bot 97(3):476–477. https://doi.org/10.1093/aob/mcj057
Xu Z, Wang H (2007) LTR-FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 35(SUPPL.2):265–268. https://doi.org/10.1093/nar/gkm286
Xu Y, Du J (2014) Young but not relatively old retrotransposons are preferentially located in gene-rich euchromatic regions in tomato (Solanum lycopersicum) plants. Plant J 80(4):582–591. https://doi.org/10.1111/tpj.12656
Xu, Z, Liu, J, Ni, W, Peng, Z, Guo, Y, Ye, W, … Du, J (2017) GrTEdb: the first web-based database of transposable elements in cotton (Gossypium raimondii). Database, 2017(1), 1–7. https://doi.org/10.1093/database/bax013
Yadav CB, Bonthala VS, Muthamilarasan M, Pandey G, Khan Y, Prasad M (2015) Genome-wide development of transposable elements-based markers in foxtail millet and construction of an integrated database. DNA Res 22(1):79–90. https://doi.org/10.1093/dnares/dsu039
Yamada, NA, Rector, LS, Tsang, P, Carr, E, Scheffer, A, Sederberg, MC, ... & Brothman, AR (2011) Visualization of fine-scale genomic structure by oligonucleotide-based high-resolution FISH. Cytogen Genome Res 132(4), 248–254. https://doi.org/10.1159/000322717
Yang, S., Zeng, K., Chen, K., Zhao, X., Wu, J., Huang, Y., … Deng, Z. (2020). Sequence evolution, abundance, and chromosomal distribution of Ty1-copia retrotransposons in the Saccharum spontaneum genome. Cytogen Genome Res 160(5), 272–282. https://doi.org/10.1159/000506222
Zhang QJ, Gao LZ (2017) Rapid and recent evolution of LTR retrotransposons drives rice genome evolution during the speciation of AA- genome Oryza species. Genes|Genomes|Gen. https://doi.org/10.1534/g3.116.037572
Zhao M, Ma J (2013) Co-evolution of plant LTR-retrotransposons and their host genomes. Protein Cell 4(7):493–501. https://doi.org/10.1007/s13238-013-3037-6
Zuccolo, A, Sebastian, A, Talag, J, Yu, Y, Kim, HR, Collura, K, … Wing, RA (2007) Transposable element distribution, abundance and role in genome size variation in the genus Oryza. BMC Evol Biol, 7, 1–15. https://doi.org/10.1186/1471-2148-7-152
Acknowledgements
The authors acknowledge Dr. Maria Virginia Sanchez-Puerta for her valuable help with phylogenetic tree construction, Dr. Carlos Llorens for his support in retroelements classification, Dr. Thomas Wicker for providing retroelement reference sequences, and to Dr. Alejandra Trenchi for sharing her experience during experiments FISH. We also appreciate the valuable language assistance of Lic. Vanesa Heredia and Dr. Hernan G. Rosli. The authors are also grateful for anonymous reviewers' and the editor Dr. Jiming Jiang's comments which improved substantially this work. This work was part of the PhD thesis of YSA, which benefited from AGENCIA and CONICET fellowship.
Funding
This work was funded by Agencia Nacional de Promoción Científica y Tecnológica, Argentina (AGENCIA), D-TEC 0008/13; PROJOVI 2015 and 2018 (Universidad Nacional de San Juan); and FONCyT, PICT-2018-01691 (AGENCIA).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Jiming Jiang
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yañez-Santos, A.M., Paz, R.C., Paz-Sepúlveda, P.B. et al. Full-length LTR retroelements in Capsicum annuum revealed a few species-specific family bursts with insertional preferences. Chromosome Res 29, 261–284 (2021). https://doi.org/10.1007/s10577-021-09663-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-021-09663-4