Abstract
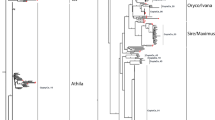
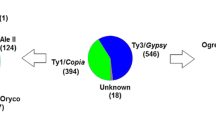
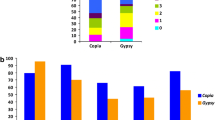
Transposable elements are the most abundant components of plant genomes and can dramatically induce genetic changes and impact genome evolution. In the recently sequenced genome of tomato (Solanum lycopersicum), the estimated fraction of elements corresponding to retrotransposons is nearly 62%. Given that tomato is one of the most important vegetable crop cultivated and consumed worldwide, understanding retrotransposon dynamics can provide insight into its evolution and domestication processes. In this study, we performed a genome-wide in silico search of full-length LTR retroelements in the tomato nuclear genome and annotated 736 full-length Gypsy and Copia retroelements. The dispersion level across the 12 chromosomes, the diversity and tissue-specific expression of those elements were estimated. Phylogenetic analysis based on the retrotranscriptase region revealed the presence of 12 major lineages of LTR retroelements in the tomato genome. We identified 97 families, of which 77 and 20 belong to the superfamilies Copia and Gypsy, respectively. Each retroelement family was characterized according to their element size, relative frequencies and insertion time. These analyses represent a valuable resource for comparative genomics within the Solanaceae, transposon-tagging and for the design of cultivar-specific molecular markers in tomato.




Similar content being viewed by others
References
Araujo PG, Casacuberta JM, Costa APP et al (2001) Retrolyc1 subfamilies defined by different U3 regulatory regions in the Lycopersicon genus. Mol Gen Genom 266:35–41
Bergman CM, Quesneville H (2007) Discovering and detecting transposable elements in genome sequences. Brief Bioinform 8(6):382–392. doi:10.1093/bib/bbm048
Beulé T, Agbessi MD, Dussert S et al (2015) Genome-wide analysis of LTR-retrotransposons in oil palm. BMC Genom 16:1–14. doi:10.1186/s12864-015-2023-1
Biémont C, Vieira C (2006) Genetics: junk DNA as an evolutionary force. Nature 443:521–524. doi:10.1038/443521a
Bowen NJ, Jordan IK, Epstein J a et al (2003) Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res 13:1984–1997. doi:10.1101/gr.1191603
Cheng X, Zhang D, Cheng Z, Keller B, Ling HQ (2009) A new family of Ty1-copia-like retrotransposons originated in the tomato genome by a recent horizontal transfer event. Genetics 181:1183–1193
Di Rienzo JA, Casanoves F, Balzarini MG et al (2017) InfoStat versión 2017. Grupo InfoStat, FCA. Universidad Nacional de Córdoba, Argentina. URL http://www.infostat.com.ar
Domingues DS, Cruz GMQ, Metcalfe CJ et al (2012) Analysis of plant LTR-retrotransposons at the fine-scale family level reveals individual molecular patterns. BMC Genom 13:137. doi:10.1186/1471-2164-13-137
Du J, Tian Z, Hans CS et al (2010) Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J 63:584–598. doi:10.1111/j.1365-313X.2010.04263.x
El Baidouri M, Panaud O (2013) Comparative genomic paleontology across plant kingdom reveals the dynamics of TE-driven genome evolution. Genome Biol Evol 5:954–965. doi:10.1093/gbe/evt025
Feschotte C, Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41:331–368. doi:10.1146/annurev.genet.40.110405.090448
Ganal MW, Lapitan NLV, Tanksley SD (1988) A molecular and cytogenetic survey of major repeated DNA sequences in tomato (Lycopersicon esculentum). Mol Gen Genet 213:262–268
Gao D, Abernathy B, Rohksar D et al (2014) Annotation and sequence diversity of transposable elements in common bean (Phaseolus vulgaris). Front Plant Sci 5:339. doi:10.3389/fpls.2014.00339
Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi:10.1093/molbev/msp259
Grandbastien MA, Spielmann A, Caboche C (1989) Tnt1, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature 337:376–380
Havecker ER, Gao X, Voytas DF (2004) The diversity of LTR retrotransposons. Genome Biol 5(6):225. doi:10.1186/gb-2004-5-6-225
Hirochika H, Okamoto H, Kakutani T (2000) Silencing of retrotransposons in Arabidopsis and reactivation by the ddm1 mutation. Plant Cell 12:357–368
Huang CRL, Burns KH, Boeke JD (2012) Active transposition in genomes. Annu Rev Genet 46:651–675. doi:10.1146/annurev-genet-110711-155616
Hua-Van A, Le Rouzic A, Boutin TS et al (2011) The struggle for life of the genome’s selfish architects. Biol Direct 6:19. doi:10.1186/1745-6150-6-19
Jiang N, Gao D, Xiao H, van der Knaap E (2009) Genome organization of the tomato sun locus and characterization of the unusual retrotransposon Rider. Plant J 60:181–193. doi:10.1111/j.1365-313X.2009.03946.x
Kim D, Langmead B, Salzberg S (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360. doi:10.1038/nmeth.3317
Krebs CJ (1999) Estimation of Survival Rates. In: Ecological Methodology. pp 499–539
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532. doi:10.1146/annurev.genet.33.1.479
Le Rouzic A, Capy P (2006) Population genetics models of competition between transposable element subfamilies. Genetics 174:785–793. doi:10.1534/genetics.105.052241
Li W, Zhang P, Fellers JP et al (2004) Sequence composition, organization, and evolution of the core Triticeae genome. Plant J 40:500–511. doi:10.1111/j.1365-313X.2004.02228.x
Llorens C, Futami R, Covelli L et al (2011) The Gypsy Database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res 39:D70–D74. doi:10.1093/nar/gkq1061
Lucas H, Feuerbach F, Grandbastien MA, Caboche M (1995) The tobacco retrotransposon Tnt1 transposes in Arabidopsis thaliana. EMBO J 14:2364–2373
Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14:860–869. doi:10.1101/gr.1466204
Manetti ME, Rossi M, Costa APP et al (2007) Radiation of the Tnt1 retrotransposon superfamily in three Solanaceae genera. BMC Evol Biol 7:34. doi:10.1186/1471-2148-7-34
Manetti ME, Rossi M, Nakabashi M et al (2009) The Tnt1 family member Retrosol copy number and structure disclose retrotransposon diversification in different Solanum species. Mol Genet Genomics 281:261–271. doi:10.1007/s00438-008-0408-4
Marcon HS, Domingues DS, Silva JC et al (2015) Transcriptionally active LTR retrotransposons in Eucalyptus genus are differentially expressed and insertionally polymorphic. BMC Plant Biol 15:198. doi:10.1186/s12870-015-0550-1
Margalef DR (1958) Information theory in ecology. Gen Syst 3:36–71
Nagaki K, Cheng Z, Ouyang S et al (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36:138–145
Park M, Jo S, Kwon J-K et al (2011) Comparative analysis of pepper and tomato reveals euchromatin expansion of pepper genome caused by differential accumulation of Ty3/Gypsy-like elements. BMC Genom 12:85. doi:10.1186/1471-2164-12-85
Parniske M, Wulff BBH, Bonnema G et al (1999) Homologues of the Cf-9 disease resistance gene (Hcr9s) are present at multiple loci on the short arm of tomato chromosome 1. Mol Plant Microbe Interact 12:93–102
Paterson AH, Bowers JE, Bruggmann R et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Paz RC, Rendina González AP, Ferrer MS, Masuelli RW (2015) Short-term hybridization activates Tnt1 and Tto1 Copia retrotransposons in wild tuber-bearing Solanum species. Plant Biol 17(4):860–869. doi:10.1111/plb.12301
Pearce SR, Pich U, Harrison G et al (1996) The Ty1-Copia group retrotransposons of Allium cepa are distributed throughout the chromosomes but are enriched in the terminal hetero- chromatin. Chromosome Res 4(5):357–364
Pereira V (2004) Insertion bias and purifying selection of retrotransposons in the Arabidopsis thaliana genome. Genome Biol 5:R79
Perez-Hormaeche J, Potet F, Beauclair L et al (2008) Invasion of the Arabidopsis genome by the tobacco retrotransposon Tnt1 is controlled by reversible transcriptional gene silencing. Plant Physiol 147:1264–1278
Picault N, Chaparro C, Piegu B et al (2009) Identification of an active LTR retrotransposon in rice. Plant J 58:754–765. doi:10.1111/j.1365-313X.2009.03813.x
Piegu B, Guyot R, Picault N et al (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16:1262–1269. doi:10.1101/gr.5290206
Pritham EJ (2009) Transposable elements and factors influencing their success in eukaryotes. J Hered 100:648–655. doi:10.1093/jhered/esp065
Pritham EJ, Putliwala T, Feschotte C (2007) Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene 390:3–17. doi:10.1016/j.gene.2006.08.008
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41(D1):D590–D596. doi:10.1093/nar/gks1219
Salazar M, González E, Casaretto J a et al (2007) The promoter of the TLC1.1 retrotransposon from Solanum chilense is activated by multiple stress-related signaling molecules. Plant Cell Rep 26:1861–1868. doi:10.1007/s00299-007-0375-y
Sanmiguel P, Bennetzen JL (1998) Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann Bot Lon 82:37–44
Schnable PS et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115
Shannon CE, Weaver W (1949) The mathematical theory of communication. University Illinois Press, Urbana
Slotkin R, Martienssen R (2007) Transposable elements and the epi- genetic regulation of the genome. Nat Rev Genet 8:272–285
Tam SM, Causse M, Garchery C et al (2007) The distribution of copia-type retrotransposons and the evolutionary history of tomato and related wild species. J Evol Biol 20:1056–1072. doi:10.1111/j.1420-9101.2007.01293.x
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599
The Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641. doi:10.1038/nature
Tian Z, Rizzon C, Du J, Zhu L, Bennetzen JL et al (2009) Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons? Genome Res 19:2221–2230
Vicient CM (2010) Transcriptional activity of transposable elements in maize. BMC Genomics 11:1–10. doi:10.1186/1471-2164-11-601
Vitte C, Estep MC, Leebens-Mack J, Bennetzen JL (2013) Young, intact and nested retrotransposons are abundant in the onion and asparagus genomes. Ann Bot 112:881–889. doi:10.1093/aob/mct155
Voytas DF, Boeke JD (2002) Ty1 and Ty5 of Saccharomyces cerevisiae. In: Craig NL et al (eds) Mobile DNA II. ASM, Washington, DC, pp 631–683
Wang Y, Tang X, Cheng Z et al (2006) Euchromatin and pericentromeric heterochromatin: comparative composition in the tomato genome. Genetics 172:2529–2540. doi:10.1534/genetics.106.055772
Wicker T, Keller B (2007) Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res 17:1072–1081. doi:10.1101/gr.6214107
Wicker T, Sabot F, Hua-Van A et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982. doi:10.1038/nrg2165
Wilhelm M, Wilhelm FX (2001) Reverse transcription of retroviruses and LTR retrotransposons. Cell Mol Life Sci 58(9):1246–1262. doi:10.1007/PL00000937
Wollrab C, Heitkam T, Holtgräwe D et al (2012) Evolutionary reshuffling in the Errantivirus lineage Elbe within the Beta vulgaris genome. Plant J 72:636–651. doi:10.1111/j.1365-313X.2012.05107.x
Xu Y, Du J (2014) Young but not relatively old retrotransposons are preferentially located in gene-rich euchromatic regions in tomato plants. Plant J 80(4):582–591. doi:10.1111/tpj.12656
Xu Z, Wang H (2007) “LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons”. Nucleic Acids Res 35(web server issue):W265–W268
Xu Z, Liu J, Ni W et al (2017) GrTEdb: the first web-based database of transposable elements in cotton (Gossypium raimondii). Database (Oxford) 2017:1–7. doi:10.1093/database/bax013
Yang T-J, Lee S, Chang S-B et al (2005) In-depth sequence analysis of the tomato chromosome 12 centromeric region: identification of a large CAA block and characterization of pericentromere retrotranposons. Chromosoma 114:103–117. doi:10.1007/s00412-005-0342-8
Yin H, Liu J, Xu Y et al (2013) TARE1, a mutated copia-like LTR retrotransposon followed by recent massive amplification in tomato. PLoS One. doi:10.1371/journal.pone.0068587
Yin H, Du J, Wu J et al (2015) Genome-wide annotation and comparative analysis of long terminal repeat retrotransposons between pear species of P. bretschneideri and P. communis. Sci Rep 5:1–15. doi:10.1038/srep17644
Acknowledgements
This work was funded by Agencia Nacional de Promoción Científica y Tecnológica, Argentina (ANPCyT), D-TEC 0008/13 and PROJOVI 2015 (Universidad Nacional de San Juan). We thank C. Llorens for his valuable help with the Gypsy Database. This work was part of the undergraduate thesis of MEK.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10709_2017_9977_MOESM1_ESM.pdf
Fig. S1. Criteria employed to define and annotate a retroelement as a full-length: A) Example of a retroelement containing both LTR sequences and all of their constitutive proteins based on a search of Conserved Domains (NCBI). B) Examples of incomplete, truncated or empty LTR retroelement that were not considered in this study (PDF 3268 KB)
10709_2017_9977_MOESM2_ESM.pdf
Fig. S2. Phylogenetic tree based on amino acid sequences of the retrotranscriptase of complete Gypsy LTR retroelements from tomato including reference sequences. Numbers above the branches represent bootstrap support values > 50% (PDF 511 KB)
10709_2017_9977_MOESM3_ESM.pdf
Fig. S3. Phylogenetic tree based on amino acid sequences of the retrotranscriptase of complete Copia LTR retroelements from tomato including reference sequences. Numbers above the branches represent bootstrap support values > 50%. (PDF 757 KB)
10709_2017_9977_MOESM4_ESM.pdf
Fig. S4. Phylogenetic tree based on amino acid sequences of the RNaseH of complete Gypsy LTR retroelements from tomato including reference sequences. Numbers above the branches represent bootstrap support values > 50% (PDF 612 KB)
10709_2017_9977_MOESM5_ESM.pdf
Fig. S5. Phylogenetic tree based on amino acid sequences of the RNaseH of complete Copia LTR retroelements from tomato including reference sequences. Numbers above the branches represent bootstrap support values > 50% (PDF 542 KB)

10709_2017_9977_MOESM6_ESM.jpg
Fig. S6. Distribution of insertion times of full-length retroelement copies of Copia (π) and Gypsy (λ) retroelements identified in tomato. Estimated insertion times were divided into bins of 100,000 yr. Frequency classes were limited each 0.5 Mya (JPG 106 KB)
Rights and permissions
About this article
Cite this article
Paz, R.C., Kozaczek, M.E., Rosli, H.G. et al. Diversity, distribution and dynamics of full-length Copia and Gypsy LTR retroelements in Solanum lycopersicum . Genetica 145, 417–430 (2017). https://doi.org/10.1007/s10709-017-9977-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-017-9977-7