Abstract
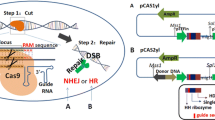
Metabolic Engineering of yeast is a critical approach to improving the production capacity of cell factories. To obtain genetically stable recombinant strains, the exogenous DNA is preferred to be integrated into the genome. Previously, we developed a Golden Gate toolkit YALIcloneNHEJ, which could be used as an efficient modular cloning toolkit for the random integration of multigene pathways through the innate non-homologous end-joining repair mechanisms of Yarrowia lipolytica. We expanded the toolkit by designing additional building blocks of homologous arms and using CRISPR technology. The reconstructed toolkit was thus entitled YALIcloneHR and designed for gene-specific knockout and integration. To verify the effectiveness of the system, the gene PEX10 was selected as the target for the knockout. This system was subsequently applied for the arachidonic acid production, and the reconstructed strain can accumulate 4.8% of arachidonic acid. The toolkit will expand gene editing technology in Y. lipolytica, which would help produce other chemicals derived from acetyl-CoA in the future.





Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Abdel-Mawgoud AM, Stephanopoulos G (2020) Improving CRISPR/Cas9-mediated genome editing efficiency in Yarrowia lipolytica using direct tRNA-sgRNA fusions. Metab Eng 62:106–115
Abed SM, Zou X, Ali AH, Jin Q, Wang X (2017) Profiling of triacylglycerol composition in arachidonic acid single cell oil from Mortierella alpina by using ultra-performance liquid chromatography-electrospray ionization-quadrupole-time-of-flight mass spectrometry. J Food Compos Anal 62:245–253
Abeln F, Chuck CJ (2021) The history, state of the art and future prospects for oleaginous yeast research. Microb Cell Fact 20(1):1–31
Bai QY, Cheng S, Zhang JL, Li MX, Cao YX, Yuan YJ (2021) Establishment of genomic library technology mediated by non-homologous end joining mechanism in Yarrowia lipolytica. Sci China Life Sci 64(12):2114–2128
Bredeweg EL, Baker SE (2021) Strain construction for intracellular metabolic pathway localization in Y. lipolytica. Yarrowia lipolytica: Methods Protoc 2307:147–157
Calderón K, Rojas-Neyra A, Carbajal-Lévano B, Luján-Valenzuela L, Ticona J, Isasi-Rivas G, Montalvan A, Criollo-Orozco M, Huaccachi-Gonzáles E, Tataje-Lavanda L (2022) A Recombinant turkey herpesvirus expressing the f protein of newcastle disease virus genotype XII generated by NHEJ-CRISPR/Cas9 and Cre-LoxP systems confers protection against genotype XII challenge in chickens. Viruses 14(4):793
Chang LL, Lu HQ, Chen HQ, Tang X, Zhao JX, Zhang H, Chen YQ, Chen W (2022) Lipid metabolism research in oleaginous fungus Mortierella alpina: current progress and future prospects. Biotechnol Adv 54:107794
Cui ZY, Zheng HH, Zhang JH, Jiang ZN, Zhu ZW, Liu XQ, Qi QS, Hou J (2021) A CRISPR/Cas9-mediated, homology-independent tool developed for targeted genome integration in Yarrowia lipolytica. Appl Environ Microb 87(6):e02666-e2720
Gemperlein K, Dietrich D, Kohlstedt M, Zipf G, Bernauer HS, Wittmann C, Wenzel SC, Muller R (2019) Polyunsaturated fatty acid production by Yarrowia lipolytica employing designed myxobacterial PUFA synthases. Nat Commun 10(1):4055
Huang YY, Jian XX, Lv YB, Nian KQ, Gao Q, Chen J, Wei LJ, Hua Q (2018) Enhanced squalene biosynthesis in Yarrowia lipolytica based on metabolically engineered acetyl-CoA metabolism. J Biotechnol 281:106–114
Ji QC, Mai J, Ding Y, Wei YJ, Ledesma-Amaro R, Ji XJ (2020) Improving the homologous recombination efficiency of Yarrowia lipolytica by grafting heterologous component from Saccharomyces cerevisiae. Metab Eng Commun 11:e00152
Jia YL, Du F, Nong FT, Li J, Huang PW, Ma W, Gu Y, Sun XM (2023) Function of the polyketide synthase domains of Schizochytrium sp. on fatty acid synthesis in Yarrowia lipolytica. J Agric Food Chem 71(5):2446–2454
Jong IS, Yu BJ, Jang JY, Jegal J, Lee JY (2018) Improving the efficiency of homologous recombination by chemical and biological approaches in Yarrowia lipolytica. PLoS ONE 13(3):e0194954
Li YW, Yang CL, Shen Q, Peng QQ, Guo Q, Nie ZK, Sun XM, Ji XJ, Shi TQ, Huang H (2021a) YALIcloneNHEJ: an efficient modular cloning toolkit for NHEJ integration of multigene pathway and terpenoid production in Yarrowia lipolytica. Front Bioeng Biotech 9:1503
Li ZJ, Wang YZ, Wang LR, Shi TQ, Sun XM, Huang H (2021b) Advanced Strategies for the Synthesis of Terpenoids in Yarrowia lipolytica. J Agri Food Chem 69a(8):2367–2381
Li MX, Zhang JL, Bai QY, Fang LX, Song H, Cao YX (2022) Non-homologous end joining-mediated insertional mutagenesis reveals a novel target for enhancing fatty alcohols production in Yarrowia lipolytica. Front Microbiol 13:898884
Liu HH, Zeng SY, Shi TQ, Ding Y, Ren LJ, Song P, Huang H, Madzak C, Ji XJ (2017a) A Yarrowia lipolytica strain engineered for arachidonic acid production counteracts metabolic burden by redirecting carbon flux towards intracellular fatty acid accumulation at the expense of organic acids secretion. Biochem Eng J 128:201–209
Liu HH, Madzak C, Sun ML, Ren LJ, Song P, Huang H, Ji XJ (2017b) Engineering Yarrowia lipolytica for arachidonic acid production through rapid assembly of metabolic pathway. Biochem Eng J 119:52–58
Liu HH, Wang C, Lu XY, Huang H, Tian Y, Ji XJ (2019) Improved production of arachidonic acid by combined pathway engineering and synthetic enzyme fusion in Yarrowia lipolytica. J Agri Food Chem 67:9851–9857
Munyangi J, Lutgen P (2020) Artemisia plants arachidonic and other polyunsaturated fatty acids. Malaria World J 11:3
Sakuradani E, Murata S, Kanamaru H, Shimizu S (2008) Functional analysis of a fatty acid elongase from arachidonic acid-producing Mortierella alpina 1S-4. Appl Microbiol Biot 81(3):497–503
Shi TQ, Gao J, Wang W J, Wang KF, Xu GQ, Huang H (2019) CRISPR/Cas9-based genome editing in the filamentous fungus Fusarium fujikuroi and its application in strain engineering for gibberellic acid production. ACS Synth Biol 8(2):445–454
Spagnuolo M, Blenner M (2021) Gene excision by dual-guide CRISPR-Cas9. Methods Mol Biol 2307:85–94
Tokuda H, Horikawa C, Nishita Y, Nakamura A, Kato T, Kaneda Y, Obata H, Rogi T, Nakai M, Shimokata H, Otsuka R (2022) The association between long-chain polyunsaturated fatty acid intake and changes in brain volumes among older community-dwelling Japanese people. Neurobiol Aging 117:179–188
Verbeke J, Beopoulos A, Nicaud JM (2013) Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnol Lett 35:571–576
Wang J, Ledesma-Amaro R, Wei Y, Ji B, Ji XJ (2020) Metabolic engineering for increased lipid accumulation in Yarrowia lipolytica–a review. Bioresour Technol 313:123707
Wei LJ, Cao X, Liu JJ, Kwak S, Jin YS, Wang W, Huaa Q (2021) Increased accumulation of squalene in engineered Yarrowia lipolytica through deletion of PEX10 and URE2. Appl Environ Microbiol 87(17):e00481-e521
Xue C, Greene EC (2021) DNA repair pathway choices in CRISPR-Cas9-mediated genome editing. Trends Genet 37(7):639–656
Xue Z, Sharpe PL, Hong SP, Yadav NS, Xie D, Short DR, Damude HG, Rupert RA, Seip JE, Wang J (2013) Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat Biotechnol 31(8):734–740
Zhou Y, Khan H, Xiao JB, Cheang WS (2021) Effects of arachidonic acid metabolites on cardiovascular health and disease. Int J Mol Sci 22(21):12029
Acknowledgements
The authors gratefully acknowledge all the members for their kind cooperation in completion of this work.
Funding
This work was supported by the National key research and development program of China (2021YFC2104300) and Natural Science Foundation of Jiangsu Province (BK20210573).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. STQ designed the experiments. SQ and LYW performed the experiments. LYW, YF, WJ, JJ, YWX, HDC and SP analyzed the data. LYW wrote the manuscript. STQ revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
This article does not contain any studies involving human or animal participants conducted by any of the authors.
Consent to participate
Written informed consent was obtained from the parents.
Informed consent
Not applicable.
Consent for publication
All authors have read the manuscript and agreed to publish.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shen, Q., Yan, F., Li, YW. et al. Expansion of YALIcloneHR toolkit for Yarrowia lipolytica combined with Golden Gate and CRISPR technology. Biotechnol Lett 46, 37–46 (2024). https://doi.org/10.1007/s10529-023-03444-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-023-03444-1