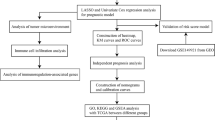
Abstract
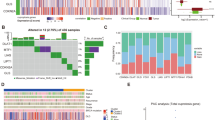
Acute myeloid leukemia (AML) is a life-threatening hematologic malignant disease with high morbidity and mortality in both adults and children. Cuproptosis, a novel mode of cell death, plays an important role in tumor development, but the functional mechanisms of cuproptosis-related genes (CRGs) in AML are unclear. The differential expression of CRGs between tumors such as AML and normal tissues in UCSC XENA, TCGA and GTEx was verified using R (version: 3.6.3). Lasso regression, Cox regression and Nomogram were used to screen for prognostic biomarkers of AML and to construct corresponding prognostic models. Kaplan–Meier analysis, ROC analysis, clinical correlation analysis, immune infiltration analysis and enrichment analysis were used to further investigate the correlation and functional mechanisms of CRGs with AML. The ceRNA regulatory network was used to identify the mRNA-miRNA-lncRNA regulatory axis. Cuproptosis-related genes LIPT1, MTF1, GLS and CDKN2A were highly expressed in AML, while FDX1, LIAS, DLD, DLAT, PDHA1, SLC31A1 and ATP7B were lowly expressed in AML. Lasso regression, Cox regression, Nomogram and calibration curve finally identified MTF1 and LIPT1 as two novel prognostic biomarkers of AML and constructed the corresponding prognostic models. In addition, all 12 CRGs had predictive power for AML, with MTF1, LIAS, SLC31A1 and CDKN2A showing more reliable results. Further analysis showed that ATP7B was closely associated with mutation types such as FLT3, NPM1, RAS and IDH1 R140 in AML, while the expression of MTF1, LIAS and ATP7B in AML was closely associated with immune infiltration. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Set Enrichment Analysis (GSEA) revealed that biological functions such as metal ion transmembrane transporter activity, haptoglobin binding and oxygen carrier activity, pathways such as interferon alpha response, coagulation, UV response DN, apoptosis, hypoxia and heme metabolism all play a role in the development of AML. The ceRNA regulatory network revealed that 6 lncRNAs such as MALAT1, interfere with MTF1 expression through 6 miRNAs such as hsa-miR-32-5p, which in turn affect the development and progression of AML. In addition, APTO-253 has the potential to become an AML-targeted drug. The cuproptosis-related genes MTF1 and LIPT1 can be used as prognostic biomarkers in AML. A total of six lncRNAs, including MALAT1, are involved in the expression and regulation of MTF1 in AML through six miRNAs such as hsa-miR-32-5p.









Similar content being viewed by others
Data Availability
The data sets analyzed during the current study are available in the TCGA (https://portal.gdc.cancer.gov/) and GTEx (https://commonfund.nih.gov/gtex).
References
Aubert L, Nandagopal N, Steinhart Z et al (2020) Copper bioavailability is a KRAS-specific vulnerability in colorectal cancer. Nat Commun 11(1):3701. https://doi.org/10.1038/s41467-020-17549-y
Bächli EB, Schaer DJ, Walter RB et al (2006) Functional expression of the CD163 scavenger receptor on acute myeloid leukemia cells of monocytic lineage. J Leukoc Biol 79(2):312–318. https://doi.org/10.1189/jlb.0605309
Bao MH, Wong CC (2021) Hypoxia, metabolic reprogramming, and drug resistance in liver cancer. Cells 10(7):1715. https://doi.org/10.3390/cells10071715
Bindea G et al (2013) Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39(4):782–795
Chen S, Chen Y, Lu J et al (2020) Bioinformatics analysis identifies key genes and pathways in acute myeloid leukemia associated with DNMT3A mutation. Biomed Res Int 23(2020):9321630. https://doi.org/10.1155/2020/9321630
Chen L, Zou W, Zhang L et al (2021) ceRNA network development and tumor-infiltrating immune cell analysis in hepatocellular carcinoma. Med Oncol 38(7):85. https://doi.org/10.1007/s12032-021-01534-6
Çiftçiler R, Haznedaroğlu İC, Sayınalp N et al (2020) The impact of early versus late platelet and neutrophil recovery after induction chemotherapy on survival outcomes of patients with acute myeloid leukemia. Turk J Haematol 37(2):116–120. https://doi.org/10.4274/tjh.galenos.2019.2019.0154
Di Nanni N, Bersanelli M, Milanesi L et al (2020) Network diffusion promotes the integrative analysis of multiple omics. Front Genet 27(11):106. https://doi.org/10.3389/fgene.2020.00106
Freshour SL, Kiwala S, Cotto KC et al (2021) Integration of the drug-gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res 49(D1):D1144–D1151. https://doi.org/10.1093/nar/gkaa1084
Gao H, Liu Y, Li K et al (2016) Hispidulin induces mitochondrial apoptosis in acute myeloid leukemia cells by targeting extracellular matrix metalloproteinase inducer. Am J Transl Res 8(2):1115–1132
Ghanem H, Tank N, Tabbara IA (2012) Prognostic implications of genetic aberrations in acute myelogenous leukemia with normal cytogenetics. Am J Hematol 87(1):69–77. https://doi.org/10.1002/ajh.22197
Gu Y, Yang R, Yang Y et al (2021) IDH1 mutation contributes to myeloid dysplasia in mice by disturbing heme biosynthesis and erythropoiesis. Blood 137(7):945–958. https://doi.org/10.1182/blood.2020007075
Gysens F, Mestdagh P, de Lavergne EDB, Maes T (2022) Unlocking the secrets of long non-coding RNAs in asthma. Thorax 77(5):514–522. https://doi.org/10.1136/thoraxjnl-2021-218359
Hänzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform 14(1):1–15
Hu X, Li J, Fu M et al (2021) The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct Target Ther 6(1):402. https://doi.org/10.1038/s41392-021-00791-1
Inui T, Wada Y, Shibuya M et al (2022) Intravenous ketogenic diet therapy for neonatal-onset pyruvate dehydrogenase complex deficiency. Brain Dev 44(3):244–248. https://doi.org/10.1016/j.braindev.2021.11.005
Ji L, Zhao G, Zhang P, Huo W, Dong P, Watari H, Jia L, Pfeffer LM, Yue J, Zheng J (2018) Knockout of MTF1 inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J Cancer 9(24):4578–4585. https://doi.org/10.7150/jca.28040
Jiang Z, Zhou X, Li R et al (2015) Whole transcriptome analysis with sequencing: methods, challenges and potential solutions. Cell Mol Life Sci 72(18):3425–3439. https://doi.org/10.1007/s00018-015-1934-y
Kahlson MA, Dixon SJ (2022) Copper-induced cell death. Science 375(6586):1231–1232. https://doi.org/10.1126/science.abo3959. (Epub 2022 Mar 17 PMID: 35298241)
Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247: 91–101; discussion 101–3, 119–28, 244–52. PMID: 12539951.
Karagkouni D, Paraskevopoulou MD, Chatzopoulos S et al (2018) DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 46(D1):D239–D245. https://doi.org/10.1093/nar/gkx1141
Kim SS, Sumner WA, Miyauchi S et al (2021) Role of B cells in responses to checkpoint blockade immunotherapy and overall survival of cancer patients. Clin Cancer Res 27(22):6075–6082. https://doi.org/10.1158/1078-0432.CCR-21-0697
Li JH, Liu S, Zhou H et al (2014) starBase v20: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. https://doi.org/10.1093/nar/gkt1248
LIU, Jianfang, et al (2018) An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173(2):400–416
Local A, Zhang H, Benbatoul KD et al (2018) APTO-253 stabilizes G-quadruplex DNA, inhibits MYC expression, and induces DNA damage in acute myeloid leukemia cells. Mol Cancer Ther 17(6):1177–1186. https://doi.org/10.1158/1535-7163.MCT-17-1209
Mayr JA, Feichtinger RG, Tort F et al (2014) Lipoic acid biosynthesis defects. J Inherit Metab Dis 37(4):553–563. https://doi.org/10.1007/s10545-014-9705-8
Nakajima W, Miyazaki K, Asano Y et al (2021) Krüppel-like factor 4 and Its activator APTO-253 induce NOXA-mediated, p53-independent apoptosis in triple-negative breast cancer cells. Genes (basel) 12(4):539. https://doi.org/10.3390/genes12040539
National Comprehensive Cancer Network NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®): Acute Myeloid Leukemia. Version 1.2016 National Comprehensive Cancer Network, Fort Washington, PA (2016) (Feb 16).
Oliveri V (2022) Selective targeting of cancer cells by copper ionophores: an overview. Front Mol Biosci 9:841814. https://doi.org/10.3389/fmolb.2022.841814
Pollyea DA, Stevens BM, Jones CL et al (2018) Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat Med 24(12):1859–1866. https://doi.org/10.1038/s41591-018-0233-1
Pomaznoy M, Ha B, Peters B (2018) GOnet: a tool for interactive Gene Ontology analysis. BMC Bioinform 19(1):470. https://doi.org/10.1186/s12859-018-2533-3
Prada-Arismendy J, Arroyave JC, Röthlisberger S (2017) Molecular biomarkers in acute myeloid leukemia. Blood Rev 31(1):63–76. https://doi.org/10.1016/j.blre.2016.08.005
Przespolewski A, Szeles A, Wang ES (2018) Advances in immunotherapy for acute myeloid leukemia. Future Oncol 14(10):963–978. https://doi.org/10.2217/fon-2017-0459
Robin X, Turck N, Hainard A et al (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform 17(12):77
Ruiz LM, Libedinsky A, Elorza AA (2021) Role of copper on mitochondrial function and metabolism. Front Mol Biosci 8:711227. https://doi.org/10.3389/fmolb.2021.711227
Seipel K, Messerli C, Wiedemann G et al (2020) MN1, FOXP1 and hsa-miR-181a-5p as prognostic markers in acute myeloid leukemia patients treated with intensive induction chemotherapy and autologous stem cell transplantation. Leuk Res 89:106296. https://doi.org/10.1016/j.leukres.2020.106296
Sha S, Si L, Wu X et al (2022) 2022 Prognostic analysis of cuproptosis-related gene in triple-negative breast cancer. Front Immunol 13:922780. https://doi.org/10.3389/fimmu.2022.922780
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Sheng XF, Hong LL, Li H et al (2021) Long non-coding RNA MALAT1 modulate cell migration, proliferation and apoptosis by sponging microRNA-146a to regulate CXCR4 expression in acute myeloid leukemia. Hematology 26(1):43–52. https://doi.org/10.1080/16078454.2020.1867781
Subramanian A, Tamayo P, Mootha VK et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102(43):15545–15550
Sun S, Yao M, Yuan L et al (2021) Long-chain non-coding RNA n337374 relieves symptoms of respiratory syncytial virus-induced asthma by inhibiting dendritic cell maturation via the CD86 and the ERK pathway. Allergol Immunopathol (madr) 49(3):100–107. https://doi.org/10.15586/aei.v49i3.85
Tomczak K, Czerwińska P, Wiznerowicz M (2015) The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (pozn) 19(1A):A68-77
Tsvetkov P, Coy S, Petrova B et al (2022) Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 375(6586):1254–1261. https://doi.org/10.1126/science.abf0529
Vivian J, Rao AA, Nothaft FA et al (2017) Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol 35(4):314–316
Vyas A, Duvvuri U, Kiselyov K (2019) Copper-dependent ATP7B up-regulation drives the resistance of TMEM16A-overexpressing head-and-neck cancer models to platinum toxicity. Biochem J 476(24):3705–3719. https://doi.org/10.1042/BCJ20190591
Widowati W, Jasaputra DK, Sumitro SB et al (2020) Effect of interleukins (IL-2, IL-15, IL-18) on receptors activation and cytotoxic activity of natural killer cells in breast cancer cell. Afr Health Sci 20(2):822–832. https://doi.org/10.4314/ahs.v20i2.36.PMID:33163049;PMCID:PMC7609126
Wu X, Sui Z, Zhang H et al (2020) Integrated analysis of lncRNA-mediated ceRNA network in lung adenocarcinoma. Front Oncol 10:554759. https://doi.org/10.3389/fonc.2020.554759
Yu G, Wang LG, Han Y et al (2012) clusterProfiler: an R package for comparing biological themes among gene clusters[J]. Omics: J Integr Biol 16(5):284–287
Funding
Tianjin Key Medical Discipline (Specialty) Construction Project, TJYX2DXK-068C, Tianjin Medical University General Hospital Clinical Research Program, 22ZYYLCCG03
Author information
Authors and Affiliations
Contributions
YL: designed and conducted the whole research. XK: revised and finalized the manuscript. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest in this work.
Ethical Approval
This paper does not involve the issue of ethics, as the TCGA is a fully open public database, all of the data was freely available online, and this study did not involve any experiments on humans or animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

10528_2023_10473_MOESM2_ESM.tiff
Supplementary file2 (TIFF 1260 KB) Functional enrichment analysis of CRGs. A–B GO analysis of CRGs. C–D KEGG analysis of CRGs. E GO and KEGG analysis of CRGs. CRGs cuproptosis-related genes; GO Gene Ontology; KEGG Kyoto Encyclopedia of Genes and Genomes
10528_2023_10473_MOESM3_ESM.tiff
Supplementary file3 (TIFF 427 KB) GSEA analysis of CRGs. A MTF1. B LIAS. C ATP7B. CRGs cuproptosis-related genes; GSEA Gene Set Enrichment Analysis
10528_2023_10473_MOESM4_ESM.tiff
Supplementary file4 (TIFF 1198 KB) Cox regression. B–C Prognostic values (OS) of MTF1 and LIPT1 expression in patients with AML evaluated by the Kaplan-Meier method (GSE2191). D–E ceRNA regulatory network. mRNAs were represented by the red diamonds, miRNAs by the blue ovals, lncRNAs by the green polygons, and targeted drug by a purple triangle
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Kan, X. Cuproptosis-Related Genes MTF1 and LIPT1 as Novel Prognostic Biomarker in Acute Myeloid Leukemia. Biochem Genet 62, 1136–1159 (2024). https://doi.org/10.1007/s10528-023-10473-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10473-y